All published articles of this journal are available on ScienceDirect.
Protein Network Analysis to Prioritize Key Genes and Pathway for Stress-Mediated Neurodegeneration
Abstract
Background:
Oxidative Stress (OS) has been implicated in the pathophysiology of many neurodegenerative diseases. OS can cause cellular damage that results in cell death due to overproduction of reactive oxygen species (ROS) that may play the crucial role in the disease progression. An impaired mechanism in correlation with reduced expression of antioxidant proteins is the very common feature among most of the age-related disorders. Various in-vitro and in-vivo studies suggest the major contribution of oxidative stress in neurodegeneration. Role of Nrf2 gene is well established as a neuroprotective gene especially in concern with stress-mediated neurodegeneration. Nrf2 is a bZIP transcription factor that forms the heterodimer with small Maf protein and transcription factor AP1 that regulates transcription by binding to ARE which coordinates the transcription of genes involved in phase II detoxification and an antioxidant defense that is used to protect the cell from oxidative stress.
Aim:
The current insilico study was attempted to prioritize key genes and pathway in stress-mediated neurodegeneration through network-based analysis.
Methods:
Protein-protein interaction network was constructed and analyzed using 63 Nrf2 regulating candidate genes obtained from NCBI database based on literature studies using STRING 10.0 database and Cytoscape v 3.6.0 software plug-in Network Analyzer. Further, the functional enrichment analysis of identified gene was done using PANTHER GENE ONTOLOGY software and DAVID tool.
Results:
Based on network topological parameter, TP53, JUN, MYC, NFE2L2, AKT1, PIK3CA & UBC were identified as the key gene in the network. Among them, TP53 gene was obtained as a super hub gene with the highest Betweenness Centrality (BC) and node degree. The functional enrichment analysis was done using PANTHER GENE ONTOLOGY software and DAVID tool reveals their significant role in neurotrophin signaling pathway, MAPK signaling pathway, cellular response to stress & in the regulation of stress.
Conclusion:
The network analysis will help in prioritizing genes in the pathway that helps in understanding the underlying mechanism of disease. Thus, further study on these genes and their biological mechanism and pathway may, therefore, provide a potential target for the treatment of stress-mediated neurodegeneration.
1. INTRODUCTION
Neurodegenerative diseases are the multifactorial disorder, which results in slow progressive loss of neurons [1, 2]. The etiology of neurodegenerative diseases is still not clear; however, it results in diverse factors such as oxidative stress, Endoplasmic Reticulum (ER) stress, mitochondrial dysfunction, accumulation of Reactive Oxygen Species (ROS), loss of mitochondrial membrane potential, and ATP depletion [3-5]. The neurological disorders, such as Alzheimer's [6], Parkinson's [7], Huntington's [8], Amyotrophic lateral sclerosis [9], Multiple Sclerosis [10], and other processes related to pathological aging [11] are associated with accumulation of abnormal protein aggregates in and around affected neurons. Oxidative stress and protein misfolding play a key role in the pathogenesis and progression of neurodegenerative diseases [12] that are characterized by fibrillar aggregates composed of misfolded proteins [13]. Neuronal death or apoptosis can be mediated by either oxidative stress or ER stress or by both at the cellular level. Thus, there have been continuing efforts to find out a target that can protect the cell against oxidative damage and have the potential to treat neurodegenerative diseases.
Decrease levels of antioxidants result in contemporizing of free radicals. In the recent years, the area of research interest focuses on the endogenous cellular anti-oxidative responses via signaling pathways involving Nrf2 [14]. Nrf2 is a bZIP belongs to the cap'n’collar family, responsible for activating transcription mediated by Antioxidant Response Element (ARE) in response to oxidative stress [15]. ARE is a cis-acting enhancer found in the 5′ flanking region of many phase II detoxification and cytoprotective genes. The ARE protects against oxidative stress, mitochondrial dysfunction, and misfolded protein. Nrf2 activity is dependent on Kelch-like ECH-associated protein 1 (Keap 1) [16], is a repressor protein that binds with Nrf2 that leads ubiquitination and promotes degradation by the proteasome [17]. On oxidation of sulfhydryl groups on specific Cysteine or Phosphorylation of Keap1, the interaction between Nrf2 and Keap is disrupted. Stabilized Nrf2 translocates into the nucleus, where it’s bind to small Maf protein and modulates transcription through ARE. The formed heterodimers bind to the ARE and coordinate the transcription of genes involved in phase II detoxification and antioxidant defense [18]. Proteins expressed by these phase II detoxifying and antioxidant genes are used to maintain redox balance as well as protect the cell from oxidative stress.
Activation of the Nrf2 pathway can boost the ability to buffer free radical generation as well as it also provides new therapeutic intervention for the treatment of neurodegenerative diseases. Nrf2 has proven activity in animal models of many neurological disorders supporting the concept of developing drug target to activate the Nrf2 pathway in the brain.
In the AD, oxidative stress has the major influence on Amyloid Precursor Protein (APP) processing and tau modification via a sequence of critical events that leads to increased brain toxicity which results in oxidative stress [19]. The cell culture studies admit the toxic effect of Aβ42 on the brain that leads to cell death via apoptosis [20]. Neurotoxic compounds, such as N-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine or its active derivative, MPPT and 6-hydroxydopamine (6-OHDA) exposure lead to oxidative stress, impair mitochondrial respiration and energy metabolism that results in neurodegeneration [21]. In the AD, blocked Nrf2 activity may lead to the neuronal dysfunction and/or loss [22]. The study on substantia nigra region of human PD brain revealed confirmation of protein glycation and nitration that leads to the oxidative damage to DNA and protein resulting from persistent oxidative trauma [23]. Comparative gene expression study of neurosphere taken from PD olfactory mucosa between PD patient and control reveals significant imbalance Nrf2-ARE pathway in PD patient [24]. In ALS, Nrf2 activators play important role in protection against oxidative stress and cell death induced by SOD1 mutant protein [24, 25]. Astrocytes expressions of Nrf2 increase the lifespan and enhance the motor neuron survival in the spinal cord of SODG93A and SODH46R/H48Q transgenic mouse model of ALS [26]. Thus increase in transcriptional activity of Nrf2, will decrease the risk and delay the aging [27, 28].
On the other hand, the activity of Nrf2 is also regulated by phosphorylation through several kinases such as phosphoinositol-3 kinase (PI3K), extracellular signal-regulated protein kinase (ERK), Protein Kinase C (PKC), and pancreas enriched kinase (PERK) [29-37] that are localized in mitochondria, a major source of intracellular oxidative stress [38]. The direct phosphorylation of Nrf2 by PERK disrupts the interaction between Nrf2 and Keap1 that leads to nuclear translocation [39]. PERK kinases activated in response to Endoplasmic Reticulum (ER) stress. It has been suggested that cell survival after ER stress is mediated by the increase in Nrf2-induced glutathione [40]. A major cause of ER stress is the deposition of unfolded proteins [41], due to two of the major pathological features of the AD (amyloid plaques and NFTs) are composed of misfolded proteins. ER stress is activated in neurons and astrocytes at some point during AD progression. Upregulation of ER stress markers has been demonstrated in postmortem brain tissues and cell-culture models of many neurodegenerative disorders, including PD, AD, Amyotrophic Lateral Sclerosis (ALS), and Huntington disease and Spinocerebellar ataxias [42]. In the AD, β-amyloid aggregation induces ER stress, altering ER and mitochondrial morphology and increasing ER and oxidative stress [43]. In PD, accumulation of misfolded proteins leads to the up-regulation of UPR. In the study, ATF6a, an ER-membrane-bound transcription factor, has been shown to be activated by protein misfolding in the ER in order to protect dopaminergic neurons from MPTP [44]. Thus, Nrf2 has proven activity in animal models of many neurological disorders supporting the concept of developing a drug target to activate the Nrf2 pathway in the brain.
Computational methods have been used widely for the identification of potential drug targets in different disease pathogenesis. Genomics-based approach combined with system biology is an efficient way to identify the set of functionally enriched genes from among the arrays of the potential gene in the network. The current insilico study is performed to identify key genes and pathway against stress-mediated neurodegeneration and their functional enrichment analysis based on protein-protein interaction network approaches.
2. METHODOLOGY
2.1. Data Collection
We mined sixty-three Nrf2 regulating gene candidates in human from NCBI database on the basis of literary studies. (Table 1) (www.ncbi.nlh.nic.gov).
| Gene Symbol | Gene ID | Gene Description |
|---|---|---|
| TP53 | 7157 | tumor protein p53 |
| BRCA1 | 672 | DNA repair associated |
| AKT1 | 207 | AKT serine/threonine kinase 1 |
| PPARG | 5468 | Peroxisome proliferator-activated receptor gamma |
| PTEN | 5728 | Phosphatase and tensin homolog |
| CDH1 | 999 | Cadherin 1 |
| MYC | 4609 | v-myc avian myelocytomatosis viral oncogene |
| CDKN1A | 1026 | Cyclin-dependent kinase inhibitor 1A |
| CDH1 | 999 | cadherin 1 |
| PIK3CA | 5290 | Phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic alpha |
| HMOX1 | 3162 | heme oxygenase 1 |
| RB1 | 614041 | RB transcriptional corepressor 1 |
| NFE2L2 | 4780 | Nuclear factor, erythroid 2 like 2 |
| GSK3B | 2932 | Glycogen synthase kinase 3 beta |
| JUN | 3725 | Jun proto-oncogene, AP-1 transcription factor |
| AHR | 196 | aryl hydrocarbon receptor |
| PRKCA | 5578 | Protein kinase C alpha |
| HDAC1 | 3065 | Histone deacetylase 1 |
| UBC | 7316 | Ubiquitin C |
| CREBBP | 1387 | CREB binding protein |
| PRKCD | 5580 | Protein kinase C delta |
| NQO1 | 1728 | NAD(P)H Quinone dehydrogenase 1 |
| FBXW11 | 23291 | F-box and WD repeat domain containing 11 |
| SQSTM1 | 8878 | Sequestosome 1 |
| SUMO1 | 7341 | Small ubiquitin-like modifier 1 |
| HDAC2 | 3066 | Histone deacetylase 2 |
| SMARCA4 | 6597 | SWI/SNF related, matrix associated, a regulator of chromatin |
| MAP2K1 | 5604 | Mitogen-activated protein kinase 1 |
| HDAC3 | 8841 | Histone deacetylase 3 |
| YY1 | 7528 | YY1 transcription factor |
| BTRC | 8945 | Beta-transducin repeat containing E3 ubiquitin protein ligase |
| CASP3 | 836 | Caspase 3 |
| KEAP1 | 9817 | kelch-like ECH associated protein 1 |
| CLTC | 1213 | Clathrin heavy chain |
| ATF4 | 468 | Activating transcription factors 4 |
| EIF2AK3 | 9451 | eukaryotic translation initiation factor 2 alpha kinase 3 |
| MAFK | 7975 | MAF bZIP transcription factor K |
| MAPK9 | 10155 | Tripartite motif containing 28 |
| TRIM28 | 5598 | Mitogen-activated protein kinase 7 |
| GADD45GIP1 | 90480 | GADD45G interacting protein 1 |
| CUL3 | 6613 | Small ubiquitin-like modifier 2 |
| SUMO2 | 5605 | Mitogen-activated protein kinase 2 |
| MAP2K2 | 5583 | Protein kinase C eta |
| PRKCH | 7832 | BTG anti-proliferation factor 2 |
| BTG2 | 6478 | Siah E3 ubiquitin protein ligase 2 |
| SIAH2 | 571 | BTB domain and CNC homolog 1 |
| BACH1 | 6047 | Ring finger protein 4 |
| RNF4 | 6942 | Transcription factor 20 |
| COPS7A | 50813 | COP9 signalosome subunit 7A |
| CASP1 ENC1 |
834 8507 |
Caspase 1 ectodermal-neural cortex 1 |
2.2. Construction of PPI Network
We made use of STRING 10.0 (Search Tool for the Retrieval of Interacting Genes), for obtaining direct and indirect human protein-protein interaction network [42]. The STRING database provides functional associations derived from sources including database, experimental, co-expression, text mining, co-occurrence, neighborhood etc with the highest confidence score. We constructed network based on the highest confidence score of 0.04, which implies that only interactions with high level of confidence in network considered as reliable PPI network. Then, the PPI constructed by STRING 10.0 was visualized by Cytoscape v 3.6.0, software used for biological network visualization, data integration and interactive network generation [43].
2.3. Topology Analysis of PPI Network
The protein-protein interaction (PPI) network was analyzed using Cytoscape Plug-in Network Analyzer 3.6.1 based on parameter including betweenness centrality (BC) and node degree. In the network, gene represents node and edges represent the interaction between nodes. The degree indicates no. of edges linked to nodes, the highest the degree of nodes represents significant biological function [44]. Betweenness centrality defines the importance of the node based on the number of shortest paths that pass through each node. In the study, the PPI network was analyzed based on these parameters.
2.4. Functional Enrichment Analysis
Functional enrichment analysis of nodes in cluster network was performed using PANTHER GENE ONTOLOGY Tool [45] and DAVID (The Database for Annotation, Visualization, and Integrated Discovery Functional Annotation Tool) [46] for understanding the biological relevance of genes in response to ER/oxidative stress. GO ontology provides common detail framework to functionally annotated gene sets. KEGG provides information about molecular interaction and network reaction as the pathway. PANTHER gene list analysis used statistical overrepresentation test to analyze functionally enriched gene network in GO Biological process. The Statistical overrepresentation test uses binomial statistical comparison. DAVID Functional Annotation tools analyze functionally enriched gene in KEGG Pathway. The functional enrichment analysis was performed based on Bonferroni correction, fold enrichment and P-values as a statistical parameter (Fig. 1).
3. RESULTS AND DISCUSSION
Network-based approach is used to construct network [47] from large gene data set that leads to the prediction of putative candidate genes, prioritizing drug targets from the network [48]. The study focuses on the inter-relationship between the various components using the PPI network and assists in the identification of novel genes associated with the disease. Studies have employed a PPI network-based approach to identify the important novel gene in response to stress-mediated neurodegeneration [49, 50]. We have taken into consideration Nrf2 regulating genes implicated in response to ER and oxidative stress-mediated neurodegeneration in human.
3.1. PPI Network
Network analysis provides information about the molecular and cellular interactions of genes/protein within the network [51]. It represents entities (nodes) and their functional co-relationships (edges). Each data type contains a different aspect of the functional role of interested genes. In the study, we investigate protein-protein interaction using available data and knowledge-based using STRING 10.0 database. PPI network constructed using 63 genes in STRING as input that results in 547 interactions between 63 nodes based on parameters including database, experimental, co-expression, text mining with the confidence score (0.007), average degree nodes 17.4 and average local clustering coefficient 0.621.
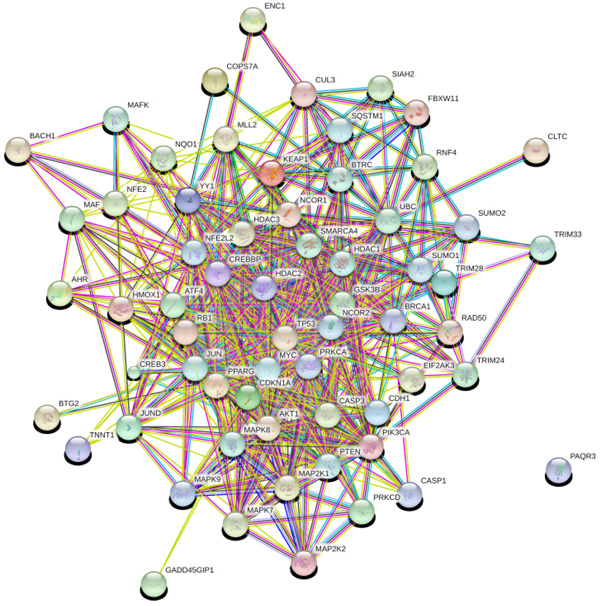
3.2. PPI Network Analysis
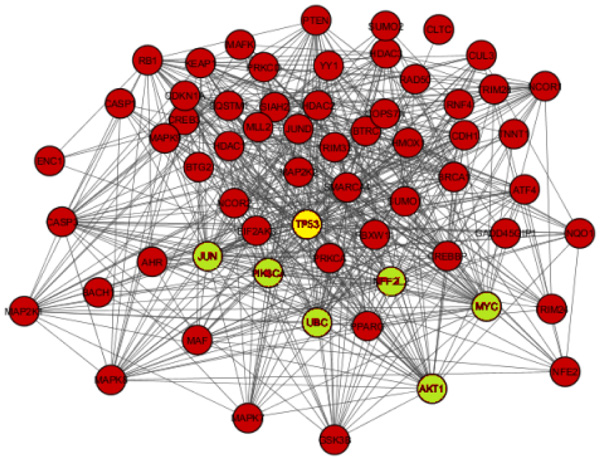
The network obtained from STRING was subsequently analyzed and visualized using Cytoscape 3.6.0 plugin Network Analyzer. In the study nodes with high degree and BC value taken as the key parameter to analyze the network Fig. (2). A cut off value for BC > 0.02 and node degree >30 consider as topological parameter for gene prioritization. TP53 (tumor protein p53), JUN (Jun proto-oncogene), UBC (Ubiquitin C), NFE2L2 (nuclear factor, erythroid 2 like 2), PIK3CA (Phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic alpha), AKT1 (AKT serine/threonine kinase 1) and MYC (v-myc avian myelocytomatosis viral oncogene) are a hub genes in network based on cut off value Table 2. Among these, TP53 is super hub gene having the highest betweenness centrality and node degree in the network (Fig. 3).
| Gene | Node Degree | Betweeness Centrality (BC) |
|---|---|---|
| TP53 | 50 | 0.13322847 |
| JUN | 43 | 0.06460235 |
| MYC | 39 | 0.0442512 |
| NFE2L2 | 35 | 005194822 |
| AKT1 | 34 | 0.02486275 |
| PIK3CA | 32 | 0.02158976 |
| UBC | 31 | 0.09984772 |
3.3. Function Annotation Analysis
For the better understanding of functional annotation of key genes in the network, we run enrichment analysis by using PANTHER GENE ONTOLOGY software and DAVID. In PANTHER Gene ontology, GO biological process reveals enrich genes including Regulation of response to stress, cellular response to stress Table 3. Pathway analysis mainly includes KEGG pathway reveals that genes are commonly enriched in neurotrophin signaling pathway, MAPK signaling pathway (Table 3).
| ID/Pathway | Term | P-value | Associated Genes |
|---|---|---|---|
| Go_0033554 | Cellular response to stress | 1.89E-06 | [TP53, UBC, AKT1, MYC, NFE2L2] |
| Go_0006950 | Response to stress | 4.79E-03 | [TP53, UBC, AKT1, MYC, NFE2L2, PIK3CA] |
| Go_ 0080134 | Regulation of response to stress | 2.85E-03 | [JUN, TP53, NFE2L2 UBC, AKT1, MYC, PIK 3CA] |
| KEGG04722 | Neurotrophin signaling pathway | 1.51E-10 | [JUN, TP53, AKT1, PIK3CA] |
| KEGG04010 | MAPK signaling Pathway | 2.6E-5 | [JUN, TP53, AKT1, MYC] |
4. DISCUSSION & CONCLUSION
Oxidative stress and protein misfolding initiate apoptotic cascades and are known to play significant roles in the pathogenesis and progression of neurodegenerative disease. Thus, targeting the Nrf2 pathway is becoming the most promising neurodegenerative therapy as it is known to induce expression of the variety of cytoprotective and detoxifying genes. Serval studies reveal the potential role of Nrf2 regulating the gene in protection from stress-induced neurodegeneration. Systems biology approaches generously contributing towards the extensive data generation which can be fruitful for better understanding as well as better cures for the stress-induced neurodegeneration.
The main aim of the study is to identify key genes and pathway against stress-mediated neurodegeneration using protein-protein interaction network analysis. The constructed PPI network consisted of total 547 interactions between 63 nodes. Based on network topology parameter i.e. Betweenness Centrality (BC >0.04) and node degree (> 30) TP53, JUN, MYC, NFE2L2, AKT1, PIK3CA & UBC were identified as the key gene in the network. Among which TP53 gene was obtained as a super hub gene with highest Betweenness Centrality (BC) and node degree. Their enrichment analysis reveals their active role in MAPK signaling pathway & neurotrophin signaling pathway.
The p53 tumor suppressor (TP53) gene regulates cell survival and death as well as cellular-redox homeostasis via modified expression of pro and anti-oxidant protein that affect mitochondrial Reactive Oxygen Species (ROS) production [52]. In the network, TP53 acts as a significant node in the intracellular control pathway, in the monitoring of cellular response to the various level of stress cell cycle arrest, replicative cell senescence, DNA repair, etc. Microglial apoptosis and microglial-induced neurotoxicity significantly reduce the treatment of microglia with p53 inhibitor pifithrin-α (PFTα) revealing the neuroprotective role of microglia p53 pathways in Alzheimer disease [53]. JUN gene is the transcription factors (AP-1) that encode the c-JUN protein that regulates pro-inflammatory cytokines, oxidative and other forms of cellular stress, and UV irradiation. The signaling pathway possesses the neural and non-neural cell that can sense oxidative stress and activate adaptive response against stress that provides strength to the anti-oxidant system. The low level of ROS, induce transcription factor nuclear factor erythroid-derived 2-related factor 2 (Nrf2) gene which is responsible for induction of many genes including NADPH Quinone oxidoreductase (NQO1), glutathione S-transferase, Heme Oxygenase-1(HO-1), ferritin, etc. [54]. Another hand, an average increase in ROS, activates AP-1 that regulates c-JUN, c-FOS etc genes. Hence, the activation of Nrf2 and AP-1 regulating genes plays the vital role in neural cell survival and protection against oxidative stress as Nrf2 form heterodimer with c-Jun that binds to ARE and regulates the transcription of many phase –II detoxifying gene in anti-oxidant defense network in the Nrf2/ARE pathway. Tp53 protein possesses an anti-oxidant property that activates the transcription of many Nrf2 coding genes and also maintains the mitochondria function in concern with ROS production [55]. In response to activation of the p53 gene by ROS, induce the Mitogen-Activated Protein Kinases (MAPKs) pathway such as p38 and c-Jun N-terminal Kinases (JNK) [56]. MAPKs (JNK & p38) pathway follow by heme- oxygenase 1(HO-1) that counteract the effect of ROS [57]. Nrf2 also interacts with and affects the Notch signaling pathway that influences cell differentiation, survival, and apoptosis. Notch signaling pathway targets the gene expression of Nrf2 coding gene and MAPKs signaling pathway via the Ras/MAPK pathway that leads to the activation of the defense system and protects against internal/external stress [58]. Thus, neuron activates the genes and induces a pathway to prevent cell death and apoptosis. The polyubiquitin gene UBC is upregulated on activation of Nrf2–Keap1 pathway under oxidative stress condition. UBC is found near the Nrf2-ARE binding site [59]. Ubc is highly demanded gene under various stress condition, thus providing valuable information underlying the mechanism of Ub in cellular defense pathway [60]. Nrf2 also regulate through an alternative mechanism including phosphorylation of Nrf2 by various protein kinases including PI3K/Akt pathway. Thus, these genes play important roles in providing strength to cellular antioxidant defenses and protect tissues from harmful damage by exerting neuroprotective, antitumor, anti-inflammatory, and antiapoptotic effects [61-65].
The network-based analysis help in prioritizing key genes network from among the vast array of potential genes in the PPI network. PPI network analysis is a key mechanism to understand all the biological processes from system biology perspective as well as also used in prediction and evaluation of corresponding treatments, providing a theoretical basis for the search of novel drug targets. In the study, we identified TP53, JUN, MYC, NFE2L2, AKT1, PIK3CA & UBC genes that play the significant role in regulating stress-mediated neurodegeneration using network-based analysis. While experimental evidence also confirms the true potential of these gene candidates in animal & human model. Further, the additional study is required to justify these finding and their potential therapeutic role in disease pathway.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
We owe deep gratitude to Dr. Aditya B. Pant, Principal Scientist, Invitro-Toxicology, CSIR-Indian Institute of Toxicology Research (IITR), Lucknow for his guidance, encouragement & support in the work. And support provided by from all those who directly or indirectly helped us in this work is gratefully acknowledged.


