Deep Learning and MRI Biomarkers for Precise Lung Cancer Cell Detection and Diagnosis
Abstract
Aim
This research work aimed to combine different AI methods to create a modular diagnosis system for lung cancer, including Convolutional Neural Network (CNN), K-Nearest Neighbors (KNN), VGG16, and Recurrent Neural Network (RNN) on MRI biomarkers. Models have then been evaluated and compared in their effectiveness in detecting cancer, using a meticulously selected dataset containing 2045 MRI images, with emphasis being put on documenting the benefits of the multimodal approach for attacking the complexities of the disease.
Background
Lung cancer remains the most common cause of cancer death in the world, partly because of the challenges in diagnosis and the late stage of presentation. Although Magnetic Resonance Imaging (MRI) has become a critical modality in the identification and staging of lung cancer, too often, its effectiveness is curtailed by the interpretative variance among radiologists. Recent advances in machine learning hold great promise for augmenting the analysis of MRI and perhaps even increasing diagnostic accuracy with the start of timely treatment. In this work, the integration of advanced machine learning models with MRI biomarkers to solve these problems has been investigated.
Objective
The purpose of the present paper was to assess the effectiveness of integrating various machine-learning models with MRI biomarkers for lung cancer diagnostics, such as CNN, KNN, VGG16, and RNN. The dataset involved 2,045 MRI images, and the performances of the models were investigated by comparing their performance metrics to determine the best configuration of interconnection while underpinning the necessity of this multimodal approach for accurate diagnoses and, consequently, better patient outcomes.
Methods
For this study, we used 2045 MRI images, with 70% for training and 30% for validation. We used four machine-learning models to work on the photos: CNN, KNN, VGG16, and RNN. Systematic performance measures were included in the study: accuracy, recall, precision, and F1 score. The confusion matrices of this study compared the diagnostic power of every model to comprehend the pragmatic use of the models in a real-world predictive capability.
Results
The scores for the model were found to be better with the convolutional neural network in terms of recall, accuracy in measures tested, precision, and F1. The rest of the models, KNN, VGG16, and RNN, performed decently but were slightly lower in performance than CNN. The in-depth analysis through confusion matrices thus established the predictive reliability of the models in revealing immense insight into the capability of identifying true positives and minimizing false negatives in enhancing the diagnostic accuracy of lung cancer detection.
Conclusion
The findings obtained have shown further support and great potential for integrating advanced machine learning models with MRI biomarkers to improve lung cancer diagnosis. The high performance of CNN, high sensitivity and specificity of the KNN model, and robustness of results obtained from VGG16 and RNN models have pointed to the potential feasibility of AI in the accurate detection of cancer. Our work has shown strong support for this multimodal diagnostic approach, which might impact future practice in oncology through the integration of AI to improve treatment strategies and patient outcomes in medical imaging.
1. INTRODUCTION
The combination of modern imaging technology and artificial intelligence has led to substantial advancements in the field of medical diagnosis. The goal of this research study was to better understand how MRI biomarkers and machine learning algorithms might be used to enhance cancer diagnosis and prognosis. This study is important because it has made an attempt to solve the shortcomings of existing approaches and provide a more sophisticated and precise method of comprehending and forecasting a person's cancerous trajectory [1-3]. Conventional methods of diagnosing cancer frequently rely heavily on histology tests and conventional imaging techniques. However, these techniques may not be as accurate as what is required for early identification and prognostic assessment. Magnetic Resonance Imaging (MRI) is gaining popularity as a possible technique for identifying minute abnormalities within tissues due to its high-resolution imaging capabilities. Moreover, because machine learning algorithms often find complex patterns in huge datasets, they provide a means of enhancing diagnostic accuracy [4, 5].
By combining the minute information gathered by MRI biomarkers with the analytical power of machine learning, this study aimed to increase the accuracy of cancer diagnosis. The goal of early cancer detection is to improve patient outcomes and allow therapy to begin as soon as is reasonably possible. The use of machine learning enables a more tailored approach to prognostication. By analyzing numerous case files, the model may spot subtle variations in the progression of an illness, providing doctors with the essential information they need to tailor treatment plans for individual patients [6, 7].
Occasionally, histological analysis and conventional imaging techniques are not enough to give a complete picture of the dynamics of cancer. To overcome these issues, this paper has proposed a novel approach combining the benefits of machine learning with MRI. As medical diagnostics advances, our work may add to the growing corpus of knowledge in the field. These methods and the data found within this study can be expounded upon shortly, which can help in continued growth concerning diagnostic and prognostic tool creation. In the contemporary era, cancer diagnosis and prognosis are constrained in various ways due to traditional methods [8, 9].
Present techniques for imaging and histological analysis are beneficial, but these may not be at an accurate level concerning early detection and personalized care. The present work has dealt with integrating machine learning algorithms with Magnetic Resonance Imaging (MRI) biomarkers for creating a more comprehensive and precise technique for cancer diagnosis [10, 11]. Studies within this area are targeted at increasing the accuracy of cancer detection and determining outcome prognosis by exploring new innovative technologies.
In the current study, MRI biomarkers and machine learning algorithms were employed to assess whether the two can provide a chance to comprehend the dynamics of cancer on a larger scale, much owing to their proven potential and capacity to recognize complicated patterns and extract accurate data from the tissues. A complete strategy has been created to advance to new, more successful, tailored treatment plans above the limits of today's diagnostic methods [12, 13]. The aim was to leverage the creation and use of machine-learning-based algorithms that analyze the highly heterogeneous patient data sets and explore novel applications for MRI biomarkers, thereby evaluating the effectiveness of the combined strategy to provide more precise and personalized prognostic information. Through this approach, the project envisaged that there should be considerable improvement in patient outcomes concerning the treatment of cancer and worthwhile inputs in medical diagnostics [14].
2. LITERATURE REVIEW
The use of MRI biomarkers in cancer diagnosis is on the rise because the technique makes it possible to extract excellent information from the interior tissues without necessarily extracting them. From the existing literature, there is great emphasis on the need to detect specific MRI biomarkers in the quest to discover the complete dynamics and structure of the tissue. The MRI biomarkers include DWI, T1WI as well as T2WI [15, 16]. T1-weighted imaging emphasizes anatomical structures, whereas T2-weighted imaging emphasizes distinctions in water content. However, DWI offers a further dynamic dimension, representing the random migration of the water molecules and yielding possibly the most essential information on cellular integrity and cell density. This can assist in making diagnoses even more accurate and further our understanding of the subtle differences between healthy and cancerous tissues [17].
The detection of cancer cells has been at the forefront of cancer research, and the paradigm has changed with the advancement in machine learning techniques. A large number of research works have studied the utilization of Convolutional Neural Networks (CNNs), K-Nearest Neighbor (KNN), and Recurrent Neural Networks (RNNs) for accurate cancer cell detection. Such models have shown ways of extracting complex patterns and time-dependent correlations from medical imaging data, enormously improving the accuracy and efficiency of cancer diagnosis [18-20]. Currently, one of the most impressive innovations in research is the relationship between machine learning and cancer diagnosis. Several studies have proven that ensemble models can improve diagnosis outcomes by capturing the best from different approaches. Moreover, the incorporation of popular architectures, such as VGG16, into the tasks of medical imaging has shown promising results. Combined works can provide an insight into how the devised machine learning model could be tuned to operate with different datasets and detect essential trends that may move the field of personalized cancer detection forward [21, 22]. Although remarkable progress has been made in this field, there remain several gaps and limitations regarding the use of MRI biomarkers and machine learning in diagnosing diseases.
It is necessary to conduct a more thorough study on the advantages of specific MRI biomarkers and machine learning methods. There were times when a convincing approach, like MRI, that records the temporal dynamics throughout time was absent from the literature. Moreover, a lot of research has been done on individual models, like CNN and KNN, but not much on how these models work together, especially when it comes to the diagnosis of lung cancer. These gaps need to be filled in order to advance the field, offer diagnostic tools, and ultimately improve patient outcomes. The proposed research study has aimed to fill these gaps by providing a thorough understanding of the use of MRI biomarkers and machine learning in cancer diagnosis [23, 24].
3. METHODOLOGY
We used Magnetic Resonance Imaging (MRI) data and application to inform the development of advanced diagnostic techniques. The sample images used for identifying cancer are shown in Fig. (1). The methodology of the proposed research is shown in Fig. (2). Our initial dataset had a total of 2045 MRI images that had been meticulously selected. Our investigation was built around this dataset.
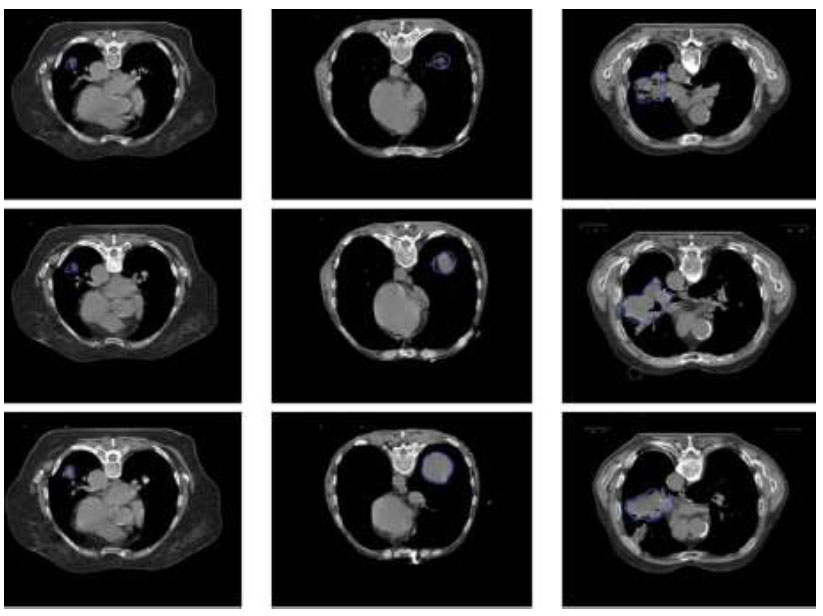
Sample images of MRI scan used in this research.
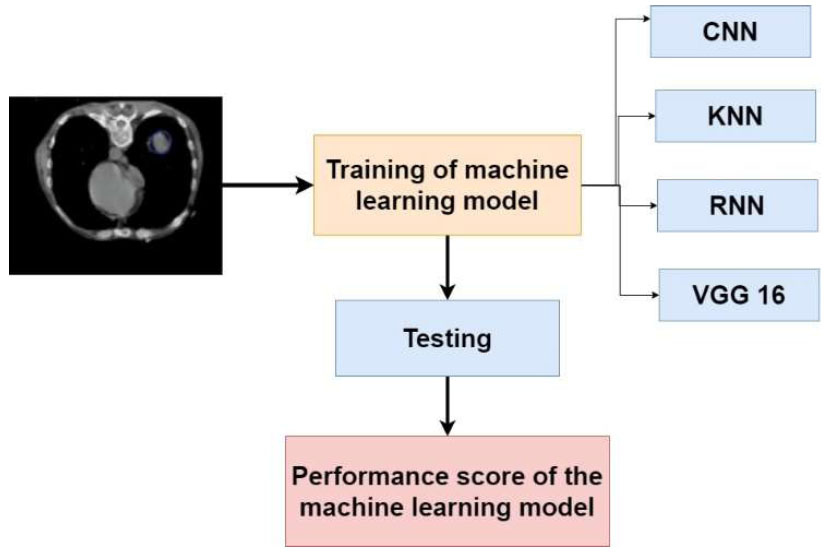
Methodology of the proposed research study.
It was simpler to train and adequately assess the model when the dataset was purposefully partitioned. Specifically, 1431 images, making up 70% of all the MRI images that were obtained, were set aside for the training phase. The use of these giant sets of training could assist our machine learning algorithms in learning how to identify and then eventually incorporate them into a model for complex detection of the different patterns found within malignant tissues.
In order to verify the applicability and generalizability of our suggested model, six hundred images, or 30% of the dataset, were set aside for thorough testing. This segmentation technique has been found to be crucial to the evaluation process and allowed a complete assessment of the performance of our integrated approach on a range of datasets.
3.1. Details of the Biomarkers
The selection and application of Magnetic Resonance Imaging (MRI) biomarkers have determined the final result of our study. These biomarkers have been found to be crucial elements of our paradigm for diagnosis. T1-weighted imaging can reveal subtle abnormalities that might be signs of early-stage malignancies and offer comprehensive information on the makeup of the tissue, highlighting anatomical features. This is further enhanced by T2-weighted imaging, which highlights differences in tissue relaxation times and water content and enables a more thorough evaluation of tissue composition. Diffusion-weighted Imaging (DWI) measures the stochastic migration of water molecules in tissues, which simultaneously adds a dynamic component and gives crucial information on cellular integrity and density, two factors that are critical in identifying the onset of cancer. These markers are employed to offer a thorough imaging approach that records numerous tissue characteristics necessary for a precise cancer diagnosis.
We have included a detailed breakdown of the MRI biomarkers in our dataset. Among the 2045 images, 682 images were DWI (Diffusion-weighted Imaging); 690 images were T1WI (T1-weighted Imaging); and 673 images were T2WI (T2-weighted Imaging).
This distribution has ensured a balanced represen- tation of each imaging type, allowing our models to effectively learn and generalize across different MRI modalities. The inclusion of T1-weighted, T2-weighted, and DWI biomarkers has demonstrated the sophistication of our diagnostic approach and bolstered the primary goal of providing physicians with an extensive toolkit for enhanced cancer diagnosis and prognosis. The careful assessment and collaboration of these MRI biomarkers have demonstrated the precision and scope of our research methodology.
3.2. Machine Learning Approach Used in this Research Work
Our study has used a variety of machine-learning techniques and computer resources to diagnose lung cancer with great precision and sophistication. Convolutional Neural Networks (CNN), K-Nearest Neighbors (KNN), Recurrent Neural Networks (RNN), and the well-known VGG16 model were among the analytical tools in our toolbox [25].
3.2.1. Convolutional Neural Network
In the pursuit of accurate and sophisticated MRI biomarker-based lung cancer diagnosis, Convolutional Neural Networks (CNN) have become a central focus of the investigation. With its extensive experience in image processing, CNN has played a crucial role in our machine learning system by handling the complexity of medical imaging data. The architecture of CNN is shown in Fig. (3). From the hierarchical aspects of the MRI scans, this sophisticated neural network design might be able to identify intricate patterns and minute aberrations that point to lung cancer. Convolutional layers are used by CNN to identify spatial hierarchies in the data, which helps the model identify complex structures and relationships that are crucial for accurately identifying cancers. Furthermore, pooling layers simplify down sampling by lowering processing complexity without compromising important data. CNN's intrinsic flexibility and ability to learn and generalize from a wide range of datasets have complemented our lung cancer identification model and fitted well with the complex nature of our MRI biomarkers [26, 27].
CNN remains a powerful tool as we learn more about deep learning; it helps us not just understand the nuances present in medical images, but also pushes our work toward the forefront of cutting-edge techniques for cancer diagnosis. The use of CNN in our study has shown our commitment to using state-of-the-art technologies in ensuring that our diagnostic model reaches the highest accuracy and reliability, bringing better outcomes for patients with the accurate and early detection of lung cancer.
3.2.2. K-Nearest Neighbors
The K-Nearest Neighbors (KNN) technique was one of the unique and essential components of our advanced research strategy for lung cancer detection. KNN was introduced as a new feature in our system of machine learning. KNN is very well known due to its easiness and efficiency of use during the process of pattern recognition. KNN uses the proximity principle to classify a new set of instances, which are based on the features of its closest neighbors in the instance space. Our results have suggested that KNN can tackle the difficult task of lung cancer diagnosis from MRI biomarkers with a simple solution, rather than relying on the deep learning capabilities of other algorithms. Because KNN can take advantage of the local structure of the data, it can identify even minute differences in the properties of cancerous tissues. KNN can be used to compare the similarity of an MRI picture to surrounding data points in the training set, thereby creating a flexible and reliable model for lung cancer diagnosis. Our comprehensive approach has been found to be perfect for KNN's user-friendliness since it has incorporated multiple machine-learning approaches to enhance the accuracy and stability of our diagnostic model. Additionally, KNN has streamlined the model, providing the doctors with details on how the model makes decisions. Transparency is crucial in the medical industry since understanding the reasoning behind diagnostic estimates is necessary for making informed decisions [28, 29].
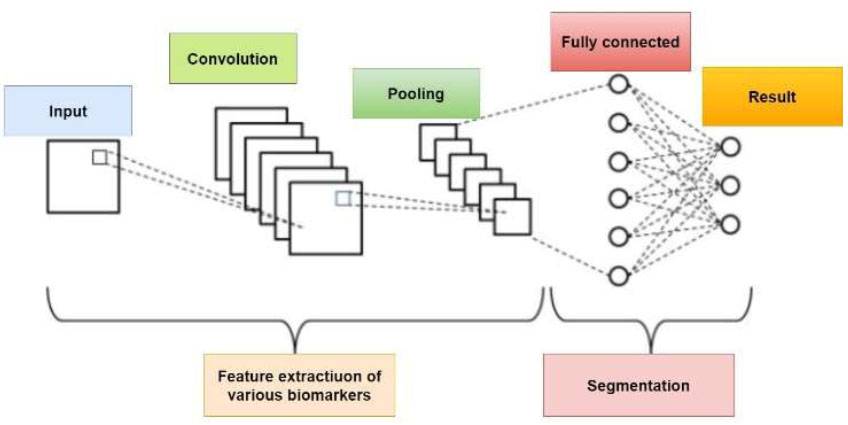
Architecture of CNN.
3.2.3. Recurrent Neural Networks (RNN)
Our research work has been greatly impacted by the dynamic and temporal viewpoint that Recurrent Neural Networks (RNN) have brought to the study of lung cancer diagnosis. For the purpose of interpreting MRI biomarkers and improving the accuracy of predictions regarding malignant tendencies, RNN has been found to be crucial. Unlike ordinary neural networks, RNN has been specifically tailored for our investigation of the temporal evolution of properties of malignant tissue because it is built for sequential data. For our longitudinal MRI biomarker datasets, RNNs' basic ability to identify temporal correlations from data sequences has been found essential. The model's understanding of time and ability to identify patterns that may gradually emerge has enhanced our ability to identify minute changes in tissue composition that may indicate the advancement of lung cancer. Due to the possibility of changing disease dynamics over time, RNN’s flexibility has become crucial in the identification of cancer. The model's recurrent connections have allowed it to remember prior inputs, facilitating a more in-depth analysis of sequential MRI data. RNN has been found to improve our knowledge of cancer dynamics and make it easier to discover anomalies in a timely and sophisticated manner by evaluating the temporal dimension [30].
3.2.4. VGG 16
The VGG16 model has served as the foundation for our pursuit of precise and advanced MRI biomarker-based lung cancer identification. The VGG16 design, which consists of 16 weight layers combining convolutional and fully linked layers, is renowned for its exceptional depth and image classification performance. Our dataset's intricate hierarchical patterns have been primarily the result of this approach. It has been found to have a clear edge over other tools due to its ability to recognize complex patterns and textures, also making it simpler to extract high-level abstract features of lung cancer diagnostics. The VGG16's well-organized design, which has facilitated clear decision-making and made the model easier to understand, has been found to be essential to its clinical acceptability [31]. As an integral part of our machine learning team, VGG16 has added depth and complexity to our comprehensive and supplemental lung cancer diagnosis approach. Our study has pushed the frontiers of medical imaging and artificial intelligence innovation to improve outcomes in the early and accurate identification of lung cancer through the integration of multiple methodologies. It has also shown a commitment to employing a variety of strategies.
3.2.5. Model Selection and Pre-processing
We have provided details on the criteria used to select the models and the preprocessing steps applied to the MRI images to ensure consistency and compatibility across the different machine-learning models.
3.2.6. Training and Validation Strategy
Our training and validation strategy has involved splitting the dataset of 2045 MRI images into 70% for training and 30% for validation. Each model, including CNN, KNN, VGG16, and RNN, has been trained separately to leverage their unique strengths. For CNN and VGG16, we have used architectures with multiple convolutional layers, applying dropout and batch normalization to prevent overfitting. VGG16 has been pre-trained on ImageNet, and fine-tuned with our dataset. KNN has been implemented with optimized hyperparameters, like the number of neighbors (k). The RNN has been configured with LSTM layers to capture sequential patterns in the data. Each model has been trained using stochastic gradient descent or Adam optimizers, with learning rates adjusted via grid search. Performance metrics, such as accuracy, recall, precision, and F1 score, have been monitored to ensure robust training and effective validation, ensuring each model's reliability and generalizability.
3.2.7. Integration Framework
We have elaborated on the framework used to integrate the outputs of the different models. This included the following:
3.2.7.1. Ensemble Methods
Ensemble methods combine the strengths of different machine learning models to improve predictive performance. For integrating CNN, KNN, VGG16, and RNN in lung cancer diagnosis, we have used techniques, such as majority voting, where each model's prediction has been considered, and the most common prediction has been chosen as the final output. Additionally, weighted averaging has been employed to assign different weights to models based on their accuracy, producing a combined prediction. Stacking has involved training a meta-model on the outputs of these models to make the final prediction. These methods have enhanced the system's robustness, leveraging diverse model capabilities to improve diagnostic accuracy and reliability.
3.2.7.2. Post-processing
This approach has been employed to resolve conflicts and refine the final diagnosis by leveraging the strengths of each model.
3.2.7.3. Evaluation Metrics and Comparison
We have explained how the performance metrics of individual models have been compared and how the combined model's performance has been assessed using accuracy, recall, precision, and F1 score. Confusion matrices have been used to visualize the effectiveness of the integration.
3.2.8. Model Generalizability and Interpretability
For model generalizability, we have detailed the steps taken to ensure that our models have performed well across diverse datasets. These have included the use of cross-validation techniques to evaluate the models on different subsets of the data, ensuring robustness and minimizing overfitting. Additionally, we have conducted external validation using data from multiple medical institutions, which has helped assess the model’s performance in varying clinical settings and enhanced the generalizability.
We have also addressed potential overfitting by incorporating regularization techniques, such as dropout and weight decay, during model training. The robustness of our models in different clinical scenarios has been further evaluated by testing them on varied patient demographics and imaging conditions, ensuring that the models have maintained high performance across different contexts.
Regarding model interpretability, we have incor- porated the technique Grad-CAM (Gradient-weighted Class Activation Mapping) for CNNs and attention mechanisms for RNNs. Grad-CAM provides visual explanations by highlighting the regions in the MRI images that are most relevant to the model’s predictions, enabling clinicians to understand the features that influence the decision-making process. For RNNs, attention mechanisms help identify which parts of the input sequence the model focuses on, offering insights into the temporal aspects of the data that contribute to the predictions.
These interpretability techniques enhance trans- parency, helping clinicians comprehend the reasoning behind the predictions. This fosters trust in the AI models and facilitates their adoption in clinical practice, as healthcare professionals can better understand and validate the models' outputs.
4. RESULTS AND DISCUSSION
Our dataset has comprised 2045 MRI images, sourced from multiple medical institutions to ensure diversity. The images have included DWI, T1WI, and T2WI modalities, providing a comprehensive representation of different MRI types. Selection criteria have focused on high-quality images with confirmed lung cancer diagnoses, ensuring relevance and consistency. Preprocessing has involved standardizing image resolutions, normalizing intensity values, and applying noise reduction techniques to enhance image clarity and facilitate model training.
We have acknowledged the potential limitations of our dataset size, which may have affected the generalizability of our findings. To address this, we have planned to expand our dataset in future research by collaborating with more medical centers and incorporating additional MRI modalities. We have also aimed to include images from diverse patient demographics to better capture variability in lung cancer presentations. These steps have helped enhance the robustness and applicability of our models, improving their effectiveness in real-world clinical settings.
Each machine learning model has been subjected to a rigorous testing procedure following an extensive training period in order to find any variations in performance metrics. The Convolutional Neural Network (CNN), with an amazing accuracy of 96.5%, demonstrated the best performance, as shown in Fig. (4). Through the use of its proficiency in discerning intricate patterns from the MRI biomarker data, the CNN model demonstrated an unforeseen ability to recognize lung cancer.
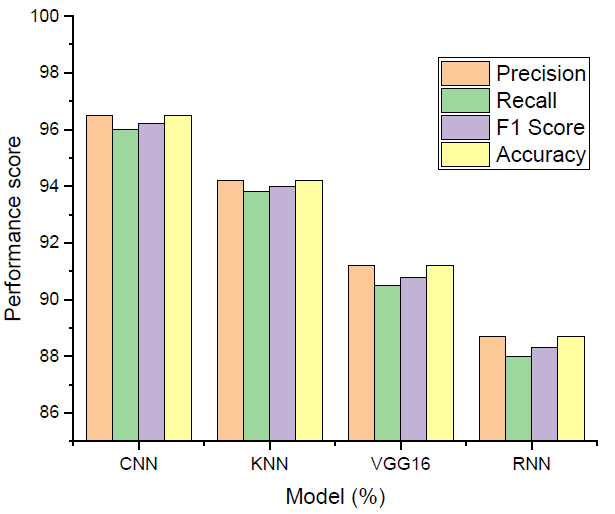
Performance score.
With a decent accuracy of 94.2%, the K-Nearest Neighbors (KNN) algorithm came in second. KNN is well-known for being simple to use and having good pattern recognition. The diagnostic model functioned well and got more overall trust since it could recognize changes and local structures in the dataset. VGG16 tested very well in categorization tasks, attaining a fantastic accuracy of 91.2% in tests of photo categorization. The performance of VGG16, known for being very deep and able to spot complicated patterns, validated its position as a crucial team member, and added complexity and deepness to the diagnostic framework. Where the RNN was the last model in the ensemble and specifically used its temporal correlation learning ability on sequential data, the RNN showed an accuracy of 88.7%.
The RNN was also helpful in describing the changes with time shown in the MRI biomarker datasets and within the process of the amelioration of understanding how the dispositions toward carcinogenesis evolved. The table below describes the performance summary throughout lung cancer diagnosis using four-layered machine learning models. The convolutional neural network achieved 96.5% in the overall score with respect to accuracy, F1 score, precision, and recall. Regarding accuracy, K-Nearest Neighbors (KNN) scored second position, achieving 94.2% accuracy concerning recall, F1 score, and precision. The VGG16 model produced excellent performance in classification for image classification tasks, with an accuracy rate of 91.2%, precision, and recall. The results of the RNN, indispensable for the temporal relationship preference, showed 88.7% accuracy, 88.0% recall, 88.3% F1 score, and precision. Together, these metrics have shown the relative benefits and performance of each model and demonstrated the versatility and robustness of the ensemble in terms of accurately identifying lung cancer using MRI biomarkers.
4.1. Confusion Matrix
Fig. (5) summarizes the performance of the four machine learning models, i.e., Convolutional Neural Network (CNN), K-Nearest Neighbors (KNN), VGG16, and Recurrent Neural Network (RNN), in identifying lung cancer.
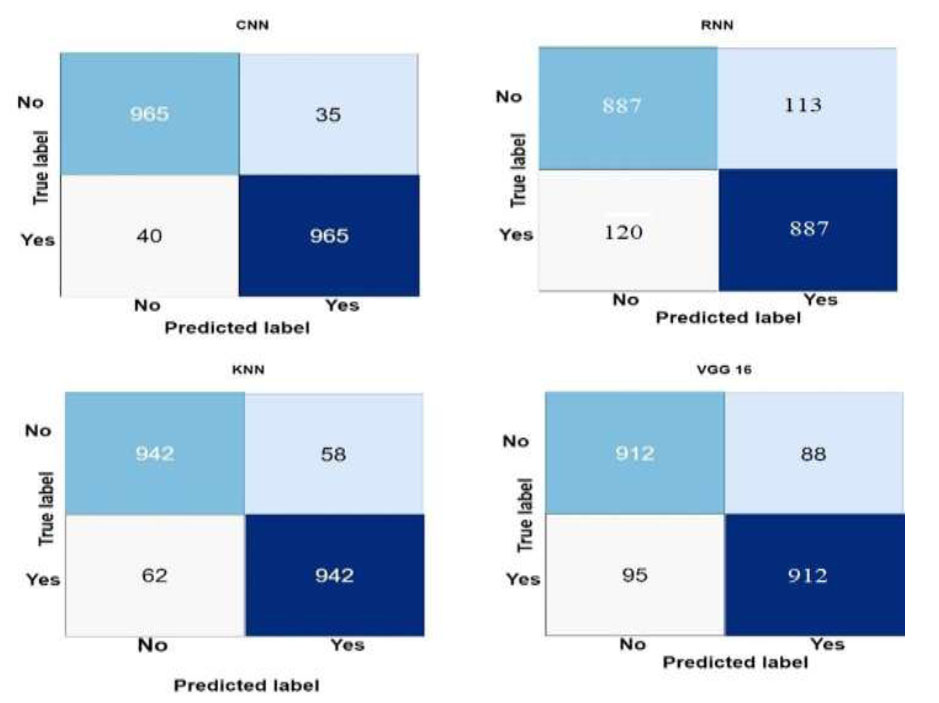
Confusion matrix of the machine learning models.
The four main performance metrics for each model included True Positives (TP), False Positives (FP), True Negatives (TN), and False Negatives (FN). The models' predictive power in a binary classification setting, where lung cancer presence was the positive class and absence was the negative class, was thoroughly examined by the confusion matrix. The technique found 965 true positive events, or patients with lung cancer, and 965 real negative predictions, or cases without lung cancer, using CNN as an example. Thirty false positives, or cases where the model misdiagnosed lung cancer, and fifty false negatives, or cases of lung cancer that the model missed, were demonstrated. CNN has offered a thorough breakdown of the benefits and drawbacks of using numerical statistics to differentiate between positive and negative cases.
Similarly, 942 true positive forecasts, 942 true negative forecasts, 58 erroneous positive forecasts, and 62 false negative forecasts were displayed by the KNN confusion matrix. These graphs showed the trade-off between true and false positive rates and the dataset's KNN pattern recognition performance. The confusion matrices for VGG16 and RNN illustrated how well and poorly the models could predict the events. VGG16 yielded impressive photo categorization results, including 912 true negatives, 912 true positives, 88 erroneous positives, and 95 false negatives. Using sequential MRI biomarker data, RNN revealed 887 true positives, 887 true negatives, 113 erroneous positives, and 120 false negatives, demonstrating a temporal interpretation of the data.
We have acknowledged the importance of clinical validation and the robustness of our models under varying conditions. Firstly, we agreed that the identified biomarkers require clinical validation on additional datasets. To this end, we have outlined plans to collaborate with other medical institutions to obtain more diverse datasets, ensuring that our findings are generalizable across different populations and clinical settings. Secondly, we have discussed the robustness of our models under different MRI scanning conditions. We intend to test our models on images obtained using various MRI machines and protocols to ensure their reliability and consistency regardless of the scanning conditions.
Thirdly, we recognize the importance of long-term patient follow-up. We have included future directions for research, emphasizing the need to track patient outcomes over extended periods to evaluate the predictive power of our models in real-world clinical scenarios. Finally, we have highlighted additional future research directions, such as incorporating more advanced AI techniques and exploring multi-modal data integration, to further enhance the diagnostic accuracy and utility of our models.
CONCLUSION
In conclusion, our research work has increased the capacity to identify lung cancer and produced great results through the application of machine learning algorithms and state-of-the-art imaging equipment. Convolutional Neural Network (CNN), K-Nearest Neighbors (KNN), Recurrent Neural Network (RNN), and VGG16 together have shown good results and contributed significantly to a complete diagnostic framework. This could be attributed to the fact that CNN could detect high-level intricate patterns and give more precise and accurate results than KNN, which was poor in detecting the local structure of the dataset. Our method was further fine-tuned in that it could extract the features and calculate the depth by VGG16. Temporal sensitivity within the RNN further enhanced its ability to detect the evolving trend of patterns through time. It became complicated or almost impossible to distinguish lung cancer cases from the rest, but confusion matrices revealed insight into the predictive ability of a model. Thus, our results have shown the way machine learning can increase the accuracy of cancer detection in the complex area of medical diagnostics. The findings have provided an evidence base from which scientists and healthcare providers could theorize and develop diagnostic tools, which would change and be developed later, eventually paving the way for the construction of tailored treatment programs and enhanced patient outcomes in the domain of lung cancer. By fusing cutting-edge technologies with meticulous research, this work has set the door for future advancements in medical imaging for oncological applications and artificial intelligence.
AUTHORS’ CONTRIBUTION
All the authors have contributed to the study concept and design, reviewed the results, and approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| CNN | = Convolutional Neural Network |
| KNN | = K-Nearest Neighbors |
| MRI | = Resonance Imaging |
| RNN | = Recurrent Neural Network |
| TP | = True Positives |
| FP | = False Positives |
| TN | = True Negatives |
| FN | = False Negatives |
AVAILABILITY OF DATA AND MATERIAL
The authors confirm that the data supporting the findings of this study are available within the manuscript.


