A Comprehensive Review of Blood Malignancy Detection in Microscopic Blood Cell Images Utilizing Complete Leukocyte Count Data
Abstract
Background
Leukemia, which is a blood cancer, is caused by the abnormal growth of white blood cells (WBCs), primarily found in the myeloid and fatty tissues of bone marrow. Microscopy is used by microbiologists and pathologists to examine the blood for the detection of leukemia. Blood cells are analyzed for morphological markers that aid in the detection and classification of leukemia. However, this method is time-consuming for malignancy prognosis and may be influenced by the clinical abilities and work experience of microbiologists.
Aims and Objectives
This research aimed to review and analyze various machine learning (ML) and deep learning (DL) approaches for the identification and categorization of different types of leukemia, particularly acute myeloid leukemia (AML) and chronic myeloid leukemia (CML), based on microscopic images of white blood cells (WBCs). It also aimed to evaluate the efficacy of various machine learning and deep learning classifiers for detecting acute and chronic myeloid leukemia and classifying different types of leukocytes.
Methods
In this study, a Support Vector Machine (SVM) classifier, representing traditional machine learning (ML) models, and a Convolutional Neural Network (CNN) classifier, based on deep learning (DL) algorithms, were employed to identify and classify myelogenous leukemia and different types of leukocytes.
Results
The algorithms utilizing the above-mentioned classifiers demonstrated significantly better performance metrics compared to other models. Conventional artificial intelligence (AI) approaches in medical image analysis have demonstrated effectiveness in accurately and reliably classifying biological images, such as microscopic blood cells, with greater precision and reliability.
Conclusion
CNNs achieved the highest accuracy, while SVMs excelled in precision among traditional methods. Combining both techniques also yielded great results. While accuracy is an important metric, it is not the only factor to consider. Overall, CNNs are more effective at detecting and classifying leukocytes and myelogenous leukaemia.
1. INTRODUCTION
The bone marrow serves as the body's central hub for producing blood cells, each with essential functions: red blood cells (RBCs) carry oxygen and remove carbon dioxide, white blood cells (WBCs) defend against infections, and platelets aid in blood clotting. Among these, WBCs are particularly critical, forming the foundation of the immune system. However, when WBC development is disrupted, it can lead to severe diseases, one of the most aggressive being Acute Lymphocytic Leukemia (ALL).
ALL is a fast-progressing form of blood cancer, most commonly diagnosed in children. It begins in the bone marrow, where immature lymphocytes (a type of WBC) begin to multiply uncontrollably, disrupting the normal production of blood cells. If left untreated, ALL can become life-threatening within a short period. Early diagnosis is vital and typically starts with microscopic examination of blood smears. However, this traditional method depends heavily on human expertise, making it time-consuming and potentially prone to misdiagnosis, especially in early or ambiguous cases.
To overcome these limitations, researchers have turned to image-based analysis and deep learning techniques for the early and accurate detection of leukemia. These methods aim to automate the identification and classification of WBCs using high-resolution blood smear images, significantly reducing subjectivity and diagnostic delays.
For instance, Awad and Aly investigated the use of object detection models, such as YOLOv8 and YOLOv11, to locate and classify leukemic cells with high accuracy [1, 2]. These models analyze features like cell shape, size, and texture to distinguish malignant WBCs from healthy ones, showing promise in assisting clinical workflows.
In a complementary approach, Chen et al. proposed DAFFNet, a Dual Attention Feature Fusion Network that leverages both spatial and channel-wise attention mechanisms to focus on the most relevant image regions for classification [3]. This attention-based method enhances the model's ability to differentiate subtle visual cues across various types of leukocytes, especially in borderline or early-stage cases.
Building on these efforts, Anand et al. developed a deep learning pipeline for the segmentation and morphological classification of leukocytes [4]. Their framework automatically annotates blood smear images and detects anomalies suggestive of leukemia, thereby accelerating diagnosis while ensuring consistency across different patient cases.
Collectively, these advancements illustrate how deep learning is transforming hematologic diagnostics. By integrating digital imaging with intelligent feature extraction and automated classification, these tools can help bridge the gap between early symptoms and confirmed diagnosis. As these AI-based systems continue to mature, they hold the potential to become invaluable decision-support tools for clinicians working in hematology and oncology.
1.1. WBC and Blood Cancer
WBCs are divided into two categories depending on the appearance of their cytoplasm. The first category comprises granulocytes, which include basophils, eosinophils, and neutrophils. Agranulocytes are the second type, which includes monocytes and lymphocytes. WBCs can also be categorized into T lymphocytes and B lymphocytes based on their functions. Different types of WBCs are shown in Fig. 1. Leukemia is a variant of blood cancer that originates in the bone marrow and leads to the production of abnormal WBCs, known as blasts or leukemia cells. These cells interfere with the circulation of normal WBCs and weaken the body's resistance [4].
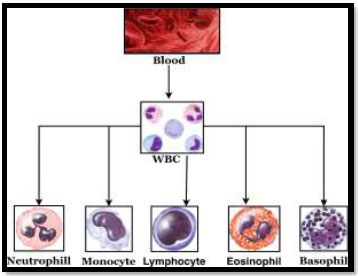
Types of WBC (granulocytes and agranulocytes).
Leukemia is classified based on the type of WBCs involved and the progression of the disease. Acute myeloid leukemia (AML) develops suddenly and progresses rapidly over days or weeks. If left untreated, acute myeloid leukemia can lead to death within a few months. On the other hand, chronic myeloid leukemia (CML) is a slow-growing blood cancer that may take several months or years to develop.
Leukemia has various forms. However, in this review, we will discuss some of the more common types. Blood lymphoblasts are the cells that initiate Acute Lymphoblastic Leukemia (ALL). Its main symptoms include a significant rise of precursor cells in the myeloid tissues and a reduced number of normal blood cells [5]. The term lymphoblast refers to aberrant lymphocytes that proliferate quickly. This kind of leukemia is more common in children, whose WBCs are not fully developed. Without any treatment, the body produces an excessive number of lymphoblasts, which can become cancerous and lead to death [6].
Acute Myelogenous Leukaemia (AML) is a type of leukemia that develops rapidly in bone marrow stem cells or granulocytes. WBCs (excluding lymphocytes and granulocytes), RBCs, and thrombocytes are all produced by myelocytes. The myeloid tissue and fatty tissue have myeloblasts, erythrocytes, or thrombocytes. Chronic Lymphocytic Leukemia (CLL) is probably the most common category of leukemia that affects adults. In this disorder, the hematopoietic cells appear mature but are often abnormal and cannot fend off the invading lymphocytes. This disease can also spread to the lymphatic system, spleen, and liver. CLL develops when numerous aberrant lymphocytes proliferate, crowding out healthy hematopoietic cells and weakening the immune system [7].
Chronic Myelogenous Leukemia (CML) is a blood cancer characterized by an increased production and accumulation of myeloid cells in the bone marrow, as shown in Fig. 2. The progression of this condition is slow and manageable. Individuals with CML can lead regular lives and are often asymptomatic.
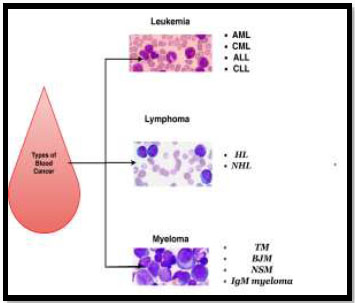
Types of blood cancer.
Machine Learning (ML) is currently one of the most popular branches of AI. It is a widely used approach for image classification, which has been a popular study area. In supervised methods, labelled data with the known outcome is accessible, and therefore, it is necessary to employ a supervised ML method. For example, the markers appear on images in WBC databases. Therefore, classifying acute leukaemia and WBCs is a supervised learning task. Traditional and DL models are widely used in the classification process of a classifier in supervised methods. Previous research on leukemia and WBC classification using supervised classification methods can be grouped into mixed classical and DL models [6].
1.2. Contributions
In this review, several blood cancer detection methods were thoroughly reviewed and critically analysed, with a focus on machine learning (ML) and deep learning (DL) approaches. Detailed discussions were conducted regarding the advantages and disadvantages of accurately detecting and categorising leukemia. The accuracy, sensitivity, and efficacy of several classifiers, including Support Vector Machines (SVMs), Convolutional Neural Networks (CNNs), and hybrid techniques, were evaluated for leukemia detection tasks. As part of the investigation, various feature extraction and image segmentation techniques were examined, with an emphasis on their practical limitations as well as their contribution to enhanced diagnostic accuracy.
Moreover, it examined challenges, including computational complexity, segmentation problems, and data quality issues. Based on the findings, recommendations and possible avenues for further research are presented.
2. LITERATURE REVIEW
A novel dataset of 500 images of peripheral blood smears with regular, acute myeloid leukemia, and acute lymphoblastic leukemia was employed in this study [14]. Nearly 1700 malignant blood cells were included in the dataset. The dimensions of images from a publicly accessible dataset were added to the collection, resulting in a heterogeneous dataset. Table 1 shows the list of databases generally used for research purposes. One of the most widely used and cost-free databases for identifying, dividing, and categorising acute leukemia is ALL-IDB [8-11].
For the automatic binary classification task, the heterogeneous dataset was employed.
Cutting-edge ML and DL techniques were employed to solve binary and three-class categorization issues. The proposed work demonstrated a binary accuracy rate of 97% by fine-tuning the fully connected convolutional layers of VGG16 and reached 98% accuracy when using DenseNet121 combined with an SVM [16]. In the case of three-class categorization, ResNet50 and the SVM achieved a 95% accuracy. The creation of the new dataset is supported by numerous experts and is expected to benefit the scientific community in advancing medical research.
Patel et al. [8] devised a technique for detecting leukemia using microscopic images of blood. For classifying WBCs, they employed histogram equalisation and the Zack method, and for detecting white cells, they applied K-means clustering. This technique was analyzed using the SVM algorithm, yielding a precision of 93.57%. The proposed computational method for identifying malignancy in blood smear images was found to be faster than the current method in terms of performance.
Raghaw et al. [17] presented CoTCoNet, a Coupled Transformer-Convolutional Neural Network model, for the classification of white blood cells in leukaemia diagnosis. The architecture combined the strengths of transformers for global feature learning and CNNs for local pattern extraction. Additionally, the model integrated a graph-based module to reconstruct cell relationships and improve classification accuracy. CoTCoNet was trained on approximately 17,000 annotated WBC images and achieved an accuracy of 98.9%. This hybrid model offered robustness against noisy data and improved interpretability. The study signifies a new trend in deep learning diagnostics by leveraging both attention mechanisms and structural connectivity information.
Krizhevsky et al. [18] and Vogado et al. [19] proposed a transfer learning-based leukemia detection technique. The feature extraction was performed using CNN (AlexNet), and feature selection was conducted using a gain ratio. On 377 photos, SVM and CNN classifiers were used. Three different datasets were used for constant reassurance, and the model achieved a classification accuracy of 99.2%.
Gayathri and Jyothi [20], developed different approaches for classifying leukocytes. Conventional feature extraction was used in the initial procedure. Classical feature extraction involves extracting features, such as territory, primary and secondary axes, and nuclei count, which are then sent to SVM algorithms and ANN classifiers for classification. For improved segmentation, Adaptive K-means clustering (AKM) was employed. Another accurate approach involves feeding a small lymphocyte image into a CNN. This method was implemented using 48 images from the database and evaluated on 36 additional images. The first approach achieved a precision of 89.47% with the SVM classifier and 92.10% with the ANN. It was found that using AKM in the segmentation stage to remove the nucleus from the algorithm yielded a more precise image for the model to interpret, and CNN classification accuracy outperformed that of SVM and ANN.
For acute myelogenous leukemia, Liu et al. [22] invented a method for separating M0 and M1 cells. The image database used for this method contained 50 photos. Otsu's Thresholding was utilised for segmentation, enabling the extraction of several significant morphological features for classification. The SMOTE was used along with RF to solve mismatches in the data. The model's categorisation precision was 89.6%. Dasariraju et al. [23] proposed an RF-based technique for identifying and categorising AML. They employed Multi-Otsu's Thresholding combined with geometric methods to segment leukocytes. Multiple features were extracted from each leukocyte image, with only the most relevant ones used during the classification stage. The dataset included 1,200 images for each type of white blood cell (WBC). The approach achieved both a prediction performance and classification accuracy of 93.44%.
By incorporating Gini importance during feature extraction and selecting highly significant features for classification, the algorithm delivered strong results in both classification and detection.
Wei et al. [24] proposed an annotation-free deep learning model for predicting genetic mutations (NPM1 and FLT3-ITD) from acute myeloid leukaemia (AML) whole-slide images (WSIs). Using a multiple-instance learning approach, the model was trained on 572 WSIs and achieved an AUC of 0.90 for NPM1 and 0.80 for FLT3-ITD mutations. This research marked a shift from morphological classification to genomic prediction directly from histopathological images. It demonstrated how computational pathology can assist in personalised treatment planning by extracting genetic information from non-annotated image regions. This innovative method reduces reliance on costly sequencing procedures and accelerates the detection of mutations.
Syed et al. [25] developed a hierarchical deep learning pipeline capable of predicting the type of leukaemia from entire microscopic blood slide images rather than just cropped or pre-annotated cell samples. Their two-stage model first classified slides into leukemic or normal types and then performed detailed subtyping using a multiclass classifier. They employed a voting mechanism over 7,255 high-powered fields (HPFs) derived from clinical data spanning 2021–2023. This innovation bridges the gap between single-cell analysis and real-world slide-level pathology, offering robust patient-level diagnosis. The study achieved high classification accuracy and demonstrated the utility of deep learning in digital pathology for leukemia diagnosis.
Adnan et al. [26] segmented cell images to provide two different outputs, one isolating the nucleus and the other capturing the cytoplasmic area. The segmentation approach achieved an accuracy of 98.33%. Yan et al. [27], presented a significant advancement in leukaemia diagnosis using a deep learning model trained on over 21,000 single-cell peripheral blood images. Their approach enabled the accurate binary and multiclass classification of acute promyelocytic leukemia (APL), non-APL, and Philadelphia chromosome-positive acute lymphoblastic leukemia (Ph+ ALL). The segmentation-enhanced residual network incorporated multigranularity training to capture diverse cell morphologies. The model achieved an F1-score of 93.2% for APL and 82.8% for non-APL cases. This model is notable for its ability to perform fine-grained leukemia typing from individual blood cells, providing a rapid and reliable diagnostic method that surpasses traditional microscopic methods. The research supports the use of automated, high-accuracy decision-making tools for haematological analysis.
Jothi et al. [9] employed the Backtracking Search Optimization Algorithm in their clustering method. The segmented nucleus images were used to extract five distinct feature types: morphological, wavelet, colour, texture, and statistical data. Feature selection is crucial in medical image processing, as it reduces both memory usage and computation time. The hybrid intelligent framework builds on the strengths of the underlying models while mitigating their limitations.
Asar and Ragab [28], introduced the FOADCNN-LDC model for the automated detection and classification of leukemia using advanced image processing and optimization techniques. The pipeline incorporated median-filter de-noising, ShuffleNetv2-based feature extraction, a convolutional autoencoder, and Falcon optimization for hyperparameter tuning. The FOADCNN-LDC model achieved an accuracy of 99.62% on publicly available leukemia datasets. The study highlighted the role of hybrid deep learning models combined with evolutionary algorithms in achieving superior diagnostic accuracy while maintaining computational efficiency. This work makes a significant contribution to real-time clinical applications and automated cancer screening solutions.
In a study, the PatternNet-fused Ensemble of Convolutional Neural Networks (PECNN), a novel technique for identifying white blood cells, was proposed [29]. The suggested architecture integrated the outputs of n randomly generated CNNs using the PatternNet ensemble approach. As it is based on randomly generated structures, the proposed technique allows for data flexibility and generality of its applications. PatternNet leverages the strengths of each participating model while remaining robust to outliers. Several experiments were carried out to demonstrate that the proposed ensemble model outperformed earlier ensemble models, even when dealing with noisy data. Moreover, the proposed architecture outperformed a more complex deep network while using significantly less computational power.
Habibzadeh et al. [30], employed a versatile thresholding method based on the Kernelized Fuzzy C-Means (KFCM) clustering technique. Instead of using a single fixed threshold, the edge estimate changed progressively based on the image content, employing what is termed adaptive thresholding. The universal edge calculation was applied to KFCM bunching and yielded a fuzzy view with fuzzy limits. Versatile and KFCM can be utilised together for clinical images and low-force images. Singhal and Singh [31], categorized blast and normal lymphocyte cells using the characteristics of the Local Binary Pattern (LBP). For the identification of ALL, LBP textural features of blood nuclei were examined. Additionally, the study compared these features with shape-based properties for classification purposes. The LBP features provided a fair level of classification accuracy. The proposed hybrid model, based on Mutual Information (MI), performed segmentation by combining the outputs of the active contour model and the Fuzzy C-Means (FCM) clustering method [29]. The accuracy, True Positive Rate (TPR), and True Negative Rate (TNR) of the blood smear images utilised in the experiment were evaluated using metrics from the AA-IDB2 database. According to the simulation results, the proposed Chronological SCA-based Deep CNN classifier achieved an accuracy rate of 98.7%.
The study identified ALL-L1, ALL-L2, and ALL-L3 as the three distinct subtypes of acute lymphoblastic leukaemia [32]. The model was found to be capable of distinguishing between normal and abnormal peripheral blood smears. Additionally, the feature values of a cancer cell and a healthy cell were determined. Using test photos contaminated with various chemicals, the model's performance was evaluated. An accuracy of 98.6% was achieved using the suggested method.
In another study, a cutting-edge full-image method was used to automatically categorize the peripheral blood smear images of multiple-nucleated acute lymphoblastic leukemia [33]. This approach sets the proposed system apart from other commonly used methods. To determine the most relevant features for the system, the authors analyzed those frequently used in existing classification systems.
White blood cells were isolated from blood samples using segmentation techniques [34]. Their color, texture, and geometric properties were then extracted and fed into various classifiers, such as Support Vector Machine (SVM), Linear Discriminant Analysis (LDA), and others, to determine whether a cell is malignant or healthy. To enhance detection accuracy, the authors compared performance with and without the use of Principal Component Analysis (PCA). If malignancy was detected, a K-Nearest Neighbors (KNN) classifier was additionally used to identify the specific type of cancer. For medical image segmentation, k-means [35] and an enhanced watershed segmentation method were developed [36]. The traditional watershed method offers the benefit of complete image separation but suffers from hypersensitivity and segmentation issues. The enhanced watershed segmentation technique utilizes automated thresholding to minimize over-segmentation and erroneous edges, resulting in segmentation maps with 92% fewer divisions compared to the typical watershed algorithm. The texture is a versatile characterisation that can be applied to a wide range of images. An architecture for a convolutional neural network (CNN) that can distinguish between blood slides, including those with ALL, AML, and healthy blood slides (HBS), was presented [37]. The model, which utilized 2,415 images from 16 datasets, yielded accuracy and precision results of 97.18% and 97.23%, respectively. The results of the proposed model were compared to those obtained using state-of-the-art methods, including those based on CNNs.
Feature extraction is a crucial step in building the patterns of the classification system, as it seeks to extract the critical description that distinguishes each class [36]. Geometrical features include radius, area, perimeter, border symmetry, shrinkage, temporal series data, eccentricity, thickness growth, and replicate elements. Texture features include homogeneity, energy, correlation, entropy contrast, and angular second moment.
Pre-trained CNNs were utilized to extract features from blood smear images and provide a distinctive visual representation [38]. PCA was chosen to select the characteristics that comprise the final descriptor after comparing various feature selection techniques. SVM, MLP, and RF were then employed to create an ensemble of classifiers that classified the images into healthy and diseased categories, achieving a 100% accuracy rate during testing.
We searched several reliable resources, including PubMed/MEDLINE, IEEE Xplore, SpringerLink, ScienceDirect, Wiley Online Library, and Google Scholar, to conduct this systematic review. We focused on research published between 2010 and 2024 that examined leukemia diagnosis using image processing and machine learning methods, particularly studies that analyzed microscopic images of blood samples. Research in the English language that met our requirements and was published in reputable conferences and peer-reviewed publications was included. Studies that were not in English, did not have full-text access, were duplicates, or only addressed molecular or genetic analysis without imaging components were not included.
Significant data, including authorship, publication year, methodology, dataset descriptions, performance metrics, and key conclusions, were extracted for each selected study. This information was combined to provide a comprehensive overview of developments in image processing-based leukemia diagnosis.
3. DETECTION AND CLASSIFICATION PROCESS FOR LEUKEMIA
3.1. Data Augmentation
Data augmentation is a technique used to enhance the diversity of the test set by creating replicas of existing data with minor modifications. Mathematical modifications are one form of data augmentation, but they are not limited to this approach. Other techniques include introducing noise, adjusting color parameters, and applying features like sharpening and blur filters to repurpose previous training samples as new data. By using data augmentation, the training dataset can be enriched without the need to collect entirely new samples, making it a cost-effective approach. Since there are already labels, data augmentation is particularly useful for supervised ML, as it eliminates the need to spend more time annotating new samples [30, 39]. (Fig. 3) presents the workflow illustrating the stages commonly involved in ML-based image analysis for leukemia detection, including data augmentation, preprocessing, segmentation, feature extraction and selection, and final classification.
Several types of AI algorithms, including contrastive learning, reinforcement learning, and generative models, contribute to the efficient augmentation of data. Data augmentation has become a standard practice in training machine learning (ML) algorithms for computer vision applications [40, 41]. Convenient methods for incorporating data augmentation into the machine learning (ML) training pipeline are readily available in advanced AI and deep learning (DL) libraries [30, 42]. However, data augmentation does not address other issues, such as perceptions in the training dataset. Additional possible issues, such as class imbalance, may need to be addressed during the data augmentation phase.
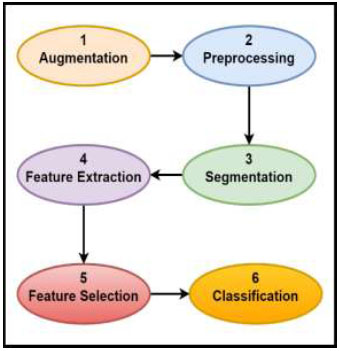
General pipeline of image-based machine learning (ML) methods for leukemia detection [19].
3.2. Preprocessing
Image enhancement is a technique used to improve the appearance of images, making them more suitable for subsequent processing. Several factors can influence the quality of visible images. According to studies, numerous methods for detecting and preparing blood images suitable for segmenting the Region of Interest (ROI) have been proposed [9, 32]. The GBR (Green, Blue, Red) colour space was converted into VHS (V: Value, S: Saturation, H: Hue) or MYKC (Magenta, Yellow, Black, and Cyan) spaces to accentuate contours for more effective ROI identification. Techniques, such as linear contrast stretching, histogram equalisation, and filters like minimum, median, and Gaussian, are used, along with normalisation and unsharp masking, to enhance the images. The approach was tested on several public WBC datasets, involving 2,551 images, and achieved a classification accuracy of 96.1%. The use of traditional neural networks in the segmentation process can further enhance accuracy. This method can be utilised as a pre-trained generative network in leukocyte subtype recognition and categorisation applications [23].
ML methods are employed to enhance the image quality. Image enhancement is a crucial and complicated stage in computer vision (CV), ML, and image processing techniques. Diagnostic systems utilize a variety of medical image types (e.g., MRI, microscope image, ultrasound image, nuclear medicine, and others). As discussed previously, many researchers have enhanced blood images by converting them to different color spaces, such as RGB to HSV or HSL, to better emphasize object properties and more effectively detect regions of interest. Low contrast, misleading backgrounds, and pepper noise are just a few issues that can degrade image quality [43]. These artifacts may emerge due to the camera and lighting conditions used to acquire the images. Various strategies have been proposed for detecting and enhancing these characteristics to make blood images acceptable for segmenting the area of interest (ROI), as shown in Fig. (4). One of the approaches for adjusting image contrast is histogram equalisation. This technique can enhance the contrast between the black backdrop and the blood images. Another approach is linear contrast, which improves and enhances the quality of blood images by boosting contrast; this technique is also known as image normalisation [44-46]. Furthermore, many proposed strategies employ a minimum filter to emphasise the lighter item, which is easy to spot during the segmentation process. However, all of the discussed techniques have drawbacks. For example, they may be susceptible to noise, helpful with low-contrast images, but unable to function effectively with blood images that contain a large percentage of noise, resulting in a loss of some clarity in the images. As a result, there is a need to search for new methods for improving image quality [45].
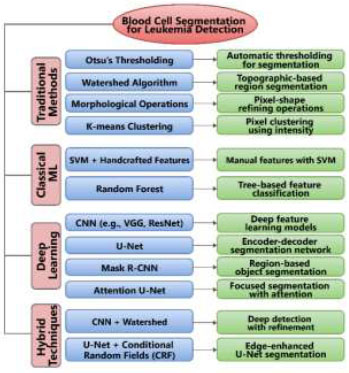
WBC segmentation techniques for leukemia detection.
3.3. Segmentation of the Image
One of the significant challenges in CV techniques is understanding digital images or gathering information from certain portions of the image. Image segmentation is the initial stage in finding an item in an image. Image segmentation [26] is a critical component in image processing, CV, and ML approaches, as it serves as a foundational step for identifying the ROI within an image. During the segmentation stage, an image is divided into several objects with similar properties based on particular criteria to obtain the region of interest. Various strategies were proposed for segmenting blast cells, associated with AML and ALL, from other components in blood smear images [47].
However, the results of these techniques have not produced optimal segmentation for complicated blood cell images. Furthermore, numerous factors made separating WBCs from the rest of the images difficult (for example, the light, contrast, and quality of medical imaging). Several features in medical blood images, such as color, shape, texture, and level of intensity, can be utilized at the segmentation step [32].
3.4. Feature Extraction
The feature extraction process captures a range of colour, texture, and morphological attributes to detect nuclei from cytoplasmic regions. Fig. (5) illustrates several methods for feature extraction in the detection of leukemia. Random Forest (RF) is employed in the classification stage, where the method achieves an accuracy of 95.86%. During preprocessing, the background and surrounding erythrocytes are removed from the white blood cell (WBC) regions [45, 48]. A binary mask is also applied at this stage to eliminate abnormal cells and reduce the risk of misclassification [19, 49]. Additionally, adjacency between leukocytes and other hematopoietic cells is reduced to improve segmentation and classification accuracy.
3.5. Classification
The classification step is one of the most significant phases in CV and ML approaches, and it is an eminent domain in this area. A collection of unstructured data can be assigned and categorised using classification [50]. The terms supervised and unsupervised classifications refer to two different types of learning approaches. In supervised classification, the collection of probable outcomes or categories is known beforehand, which is used to train the models. On the other hand, in unsupervised classification, the set of categories is unknown beforehand, and the model attempts to discover underlying patterns or groupings within the data. As shown in Fig. (4), classifiers such as Multilayer Perceptron (MLP), Support Vector Machine (SVM), Random Forest (RF), Artificial Neural Network (ANN) [45, 48], K-Nearest Neighbor (KNN), Naive Bayes (NB), and Hybrid approaches are commonly applied in supervised classification tasks [46]. Once features have been extracted from the segmented image, the next stage is to identify and classify the object type, as illustrated in Table 2 [51-54].
3.5.1. DL Approach for Leukemia Classification Methods
In this study, we also reviewed previous research that utilized Deep Neural Network (DNN) approaches for diagnosing and categorizing acute blood leukaemia. Unlike traditional models that require human feature extraction, DNN approaches efficiently perform this task automatically. As a result, DNN approaches are preferred for identifying and classifying acute leukaemia [55]. The DNN can now operate end-to-end due to the autonomous feature extraction stage, which reduces the cost of feature extraction and dimensionality reduction, as shown in Table (3). Examples of these algorithms include Deep Belief Networks (DBN), CNN [46], LSTM, RfNN, Deep
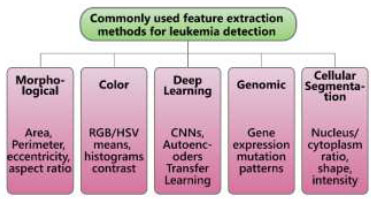
Methods for feature extraction for detection of leukemia.
| Ref. | Properties Extraction Methods | Models used | Accuracy (%) |
|---|---|---|---|
| [14] | CNN | VGG19, DenseNet121, ResNet50, SVM | 97 |
| [19] | Deep features | SVM | 99.2 |
| [17] | CNN + Transformer + Graph Reconstruction | CoTCoNet | 98.94 |
| [51] | ResNet101V2, VGG19, InceptionV3, InceptionResNetV2 + LIME | Transfer Learning with XAI | 98.38 |
| [52] | GoogleNet Features + PSO + PCA | Bayesian-optimized SVM + Subspace Ensemble (SDEL) | 97.4 |
| [21] | Transfer Learning (ResNet50) + Grad-CAM | Deep Learning Classifier with Explainable AI (XAI) | 96.81 |
| [53] | ResNet-50V2 + Genetic Algorithm (GA) | CNN with GA for Hyperparameter Tuning | 98.46 |
| [54] | EfficientNetV2M + Bayesian Optimization | CNN (EfficientNetV2M) | 91.37 |
| Ref. | Year | Feature Extraction Method | Models Used |
Accuracy (%) |
|---|---|---|---|---|
| [58] | 2025 | CNN + Transformer-based contextual learning | CoTCoNet (Transformer + CNN) | 98.9 |
| [59] | 2024 | Convolutional feature fusion + Falcon optimization | FOADCNN-LDC (Hybrid CNN + Optimized LDC) | 99.62 |
| [14] | 2022 | Deep CNN features from heterogeneous data | CNN + SVM / CNN + RF (hybrid classification) | 97 |
| [8] | 2021 | AlexNet (Deep CNN) feature extraction |
SVM (ML classifier on DL features) |
93.57 |
| [40] | 2019 | CNN features from microscopic images | D-CNN (Deep Convolutional Neural Network) | 88, for binary classification (ALL vs. healthy) |
| [39] | 2020 | CNN features | LD, DT, and K-NN (ML classifiers) |
100 |
| [57] | 2019 | Deep neural network features | SVM + NN (stacked ML and DL) | 98.8 |
Autoencoders (AE), Generative Adversarial Networks (GAN), Restricted Boltzmann Machines (RBM), and others. Ahmed et al. [40] introduced a novel approach based on neural networks for white blood cell identification, called WBCsNet. The approach utilized deep reactivation characteristics and fine-tuning of preexisting deep neural networks through transfer learning strategies [56]. Several pre-trained networks were leveraged through deep feature extraction and integrated into WBCsNet. An SVM was employed during the classification phase. The algorithm was tested on various public WBC datasets comprising 2,551 images and achieved a classification accuracy of 96.1%. The inclusion of convolutional neural networks in the segmentation stage can ensure better system accuracy. The approach can be utilized as a pre-trained model network in leukocyte subtype recognition and classification applications [6].
3.5.2. Evaluation Metrics: Precision, Recall, Specificity, and F1 Score
Evaluation metrics, such as precision, recall, specificity, and F1 score, are the classification parameters that provide a more comprehensive view of a model’s diagnostic effectiveness. They can be calculated using the following equations (1-5):
| Precision = TPR/(FPR+TPR) | (1) |
| Recall (Sensitivity) = TPR /(FNR+TPR) | (2) |
| Specificity = TNR /(FPR+TNR) | (3) |
| Accuracy = (TNR+TPR) /(FP+FN+TNR+TPR) | (4) |
| F-Measure = 2 (RecallPrecision)/(Recall+Precision) | (5) |
3.5.3. Comparison between Previous Models for the Detection of Leukaemia
While evaluating various leukaemia detection technologies, each was found to have significant shortcomings. The VGG16 model, paired with the SVM model, produced strong results, with an accuracy of 97% [14], suggesting its dependability in identifying leukaemia cells. However, due to its intricacy, this strategy may be challenging to implement in clinical practice. The CNN model utilising AlexNet demonstrated an extraordinary accuracy of 100% [57], as shown in Table (4). However, such perfect results are generally obtained under highly controlled settings, thereby limiting their real-world applicability. Excellent recall (99.55%), high precision (93.43%), and good accuracy (96.11%) were all attained using the solo SVM classifier [49]. However, it may struggle when presented with noisy or irrelevant data, as its effectiveness largely depends on the selection of high-quality, relevant features. In a similar vein, the hybrid CNN and SVM method, which required accurate feature extraction, also performed well, achieving 96.15% accuracy [6]. Therefore, despite the advantages of each approach, selecting the appropriate classifier for clinical use requires careful consideration of the computational needs, practical viability, and data quality.
| Ref. | Year | Classifier Employed | Precision (%) | Recall (%) | Specificity (%) | Accuracy (%) | F1-score (%) |
|---|---|---|---|---|---|---|---|
| [58] | 2025 | CoTCoNet (Transformer + CNN) | 97.8 | 98.4 | 96.9 | 98.9 | 98.1 |
| [59] | 2024 | FOADCNN + SVM | 99.4 | 98.6 | 97.9 | 99.62 | 99.0 |
| [14] | 2022 | VGG16+SVM | 85 | 94 | 84 | 97 | 89 |
| [8] | 2021 | CNN, SVM, Alex-net | 92.85 | 92.3 | 96.15 | 96.29 | |
| [57] | 2020 | CNN(AlexNet) | 100 | 100 | 100 | 100 | - |
| [49] | 2020 | SVM | 93.43 | 99.55 | 92.92 | 96.11 | 96.22 |
Data enhancement, visual preprocessing, segmentation, feature extraction, part selection, and categorization are all processes involved in identifying and categorizing leukemia and leukocytes. Focusing on the categorisation stage, we examined existing research in this area. Based on the classifier, we categorized the current classification techniques into three categories: classic, hybrid, and Deep Neural Networks (DNN), and analyzed their precision [35]. We also summarized the current advances in detecting and classifying ALL using deep and machine learning methods. We then analysed existing segmentation, feature extraction, and classification algorithms for efficient ALL detection. Unsupervised machine learning techniques were found to be preferred for segmentation tasks, while supervised methods were favored for classification. Deep learning, especially transfer learning, is the preferred method for detecting and classifying ALL due to its superior performance in limited datasets.
According to the survey, SVM exhibited the highest precision among other classical techniques, while CNN achieved the highest accuracy among DNN methods. Hybrid approaches that combine SVM and CNN also demonstrated excellent accuracy. Besides accuracy, other metrics also need to be considered in classification problems. In this study, various performance measures reported in prior research were also explored. It was found that works employing CNN classifiers outperformed other approaches for categorization tasks. Consequently, algorithms that utilize multiple CNN techniques for identifying and classifying myelogenous leukemia and leukocytes were found to be more successful and precise compared to those that do not. Existing methods have certain limitations; for example, the evaluated studies often used datasets with varying image resolutions, sizes, and quality. As a result, it is difficult to assess the performance of each classifier independently. Although several models demonstrated good accuracy, their testing was conducted on well-controlled datasets. This raises the question of whether these techniques would perform equally well in real-world clinical settings. SVM and other classifiers primarily rely on selecting the correct features, and accuracy can be severely compromised by noisy data or improper feature selection, which is a significant drawback. This study highlights the importance of utilizing advanced Deep Neural Networks in medical image analysis and cancer classification, which can lead to enhanced diagnostic capabilities and improved patient outcomes. As these approaches are often tested on specific, controlled datasets, their ability to generalize the findings may be limited. Furthermore, any biases in these datasets, including unequal class distribution or a lack of diversity, may affect model performance and compromise the dependability of the models when widely used in clinical practice.
CONCLUSION
Cancer has become a serious illness that affects individuals worldwide. Leukaemia, which occurs in both acute and chronic forms, can affect men and women of all ages. Acute myelogenous leukaemia (AML), a particularly aggressive form of leukaemia, is associated with a higher mortality rate. This review presents a computational approach based on leukocyte analysis to comprehensively examine each step involved in the detection and classification of acute leukaemia. The survey revealed that among deep learning methods, Convolutional Neural Networks (CNNs) achieved the highest classification accuracy. In contrast, among traditional machine learning techniques, Support Vector Machines (SVMs) demonstrated the greatest precision. Hybrid models that integrate CNNs and SVMs also yielded high accuracy. Although accuracy is a critical metric in classification problems, it is not the sole criterion for evaluation. This study also explored other performance measures discussed in previous research. CNN-based classifiers consistently outperformed alternative approaches in leukaemia classification tasks. Specifically, CNN models, such as AlexNet, achieved the highest accuracy (100%), followed by hybrid models like VGG16+SVM (97%), and traditional SVM classifiers (approximately 96%), which, although sensitive to feature quality, remained effective. The classifiers were assessed using standard evaluation metrics.
AUTHORS’ CONTRIBUTIONS
The authors confirm their contributions to the paper as follows: study concept and design: DG; conceptualization: PS; investigation: SA; visualization: PKS; drafting manuscript: YL and MD. All authors reviewed the results and approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| AML | = Acute Myeloid Leukemia |
| CML | = Chronic Myeloid Leukemia |
| WBCs | = White Blood Cells |
| SVM | = Support Vector Machine |
| ML | = Machine Learning |
| CNN | = Convolutional Neural Network |
| DL | = Deep Learning |
| AI | = Artificial Intelligence |
| ANNs | = Artificial Neural Networks |
| RBCs | = Red Blood Cells |
| ALL | = Acute Lymphocytic Leukemia |
| CLL | = Chronic Lymphocytic Leukemia |
| CML | = Chronic Myelogenous Leukemia |
AVAILABILITY OF DATA AND MATERIALS
The authors confirm that the data supporting the findings of this research are available within the article.
CONFLICT OF INTEREST
Dr. Salman Akhtar is the associate editorial board member of The Open Bioinformatics Journal.
ACKNOWLEDGEMENTS
Declared none.


