Virtual Screening and Molecular Simulation of Drug Candidates Targeting the Human SLC4A4 Protein for Colorectal Cancer Therapy
Abstract
Background
Colorectal cancer is a highly complex disease that continues to rise in prevalence, posing significant difficulties in its management and treatment outcomes. Reduced expression of the SLC4A4 gene has been identified as a critical factor in driving tumor development and poor clinical prognosis in CRC. This reduction disrupts several key cellular mechanisms, including cellular proliferation, programmed cell death, and metastasis, largely by altering pH regulation within the cells. Given its influence on tumor behavior and patient survival, SLC4A4 expression levels could serve as a reliable prognostic marker in colorectal cancer.
Methods
The CryoEM structure of SLC4A4, retrieved from the Protein Data Bank (PDB), was refined using SwissModel and subjected to virtual drug screening via the DrugRep web server. This screening employed a database comprising FDA-approved drugs.
Results
Nilotinib emerged as the most potent inhibitor following further validation through redocking using CB-DOCK2, ranking it highest among the top three results from DrugRep. The docking analysis yielded a score of −11.2 kcal/mol for Nilotinib. A 50-nanosecond molecular dynamics simulation further validated these findings, revealing that Nilotinib formed robust interactions with the SLC4A4 protein. The simulation yielded consistent results, with a root mean square deviation of around 0.30 nm, a root mean square fluctuation of near 0.5 nm, a compact radius of gyration between 3.8 and 4.0 nm, and stable solvent-accessible surface area profiles. These findings confirmed that the drug-protein complex maintained structural stability throughout the simulation.
Discussion
The computational findings suggest that Nilotinib binds effectively and stably to SLC4A4, indicating its potential to modulate the function of this protein in CRC. Its established safety profile as an FDA-approved drug supports the feasibility of drug repurposing. These results also reinforce the utility of integrating structure-based drug screening with molecular dynamics simulations to identify novel therapeutic agents for cancer.
Conclusion
Nilotinib holds significant potential as a therapeutic agent for colorectal cancer and warrants further experimental investigation to validate its effectiveness.
1. INTRODUCTION
Colorectal cancer (CRC) is among the most common and lethal malignancies worldwide, leading to an increasing percentage in cancer-related disease incidence and patient deaths [1, 2]. In this new genomic era, the knowledge of CRC has dramatically changed, revealing a layered genome that orchestrates tumor growth and biology [3]. Despite advancements in the diagnosis and treatment of colon cancer, the 5-year survival rate for advanced stages remains alarmingly low, at less than 30%, primarily due to recurrence and metastasis following surgery. Therefore, it is imperative to delve deeper into the mechanisms driving colon cancer progression and to identify new biomarkers that can enhance individualized treatment strategies and prognostic evaluations for patients [4]. Growing evidence indicates that abnormal metabolism is a pervasive feature of tumor development, significantly influencing tumor progression and resistance to therapy [5-7].
Pyruvate, the final product of glycolysis, plays a critical role in both cellular anabolism and catabolism. Studies have demonstrated that metabolic dysregulation in tumor cells is linked not only to the silencing or activation of various signaling pathways but also to the regulation of driver genes, such as those involved in partial epithelial-mesenchymal transition (p-EMT) [8-11]. Furthermore, tumor metabolites are implicated in essential cellular functions like lipid uptake, synthesis, and hydrolysis, as well as in the biological behavior of malignant tumors. Thus, exploring genes associated with pyruvate metabolism may provide valuable insights for the development of targeted tumor therapies [12, 13].
The study of gene expression patterns in various diseases is critical for understanding underlying molecular mechanisms and identifying potential therapeutic targets [14]. The Expression Atlas, a comprehensive resource providing information on gene and protein expression across multiple species and conditions, is invaluable for such analyses. This resource allows researchers to investigate the expression levels of specific genes in various tissues, cell types, and disease states [15].
One gene that has garnered significant attention in CRC research is the solute carrier family 4 member 4 (SLC4A4) gene [16]. Recent studies have further demonstrated that SLC4A4 downregulation is strongly associated with Epithelial-Mesenchymal Transition (EMT) and increased metastasis in CRC [17]. Moreover, SLC4A4 expression has been correlated with immune cell infiltration (CD8+ T cells, dendritic cells, and NK cells), underscoring its role in shaping the tumor microenvironment [18]. Cappellesso et al. (2022) highlighted that targeting SLC4A4 can overcome immunotherapy resistance, reinforcing its translational significance as both a prognostic biomarker and therapeutic target [19]. These findings highlight SLC4A4 not only as a pH regulator but also as a tumor suppressor and potential therapeutic target.
SLC4A4 encodes a sodium bicarbonate cotransporter, which plays a crucial role in maintaining cellular pH balance [20, 21]. Aberrant expression of SLC4A4 has been implicated in various cancers, including CRC [22]. The regulation of pH within the tumor microenvironment is essential for cancer cell survival and proliferation, making SLC4A4 a compelling target for therapeutic intervention [19, 23]. The ability to disrupt SLC4A4 function and alter intracellular pH levels could impede the growth and metastasis of CRC cells, offering a novel approach to treatment [24]. Previous studies have suggested that deregulation of ion transporters, including those in the SLC family, can influence cancer cell proliferation, invasion, and metastasis [25, 26]. SLC4A4, functioning as a tumor suppressor gene, experiences notable downregulation and shows a positive correlation with microsatellite instability [18].
In the effort to develop effective CRC treatments, drug repurposing has emerged as a promising strategy [27]. This involves identifying existing FDA-approved drugs that can be repurposed to target new molecular pathways implicated in CRC [28]. Virtual screening of drug libraries against the refined structure of SLC4A4 can expedite the discovery of potential inhibitors [29].
By leveraging the comprehensive genetic and functional insights provided by Expression Atlas, researchers can identify novel therapeutic targets and accelerate the development of effective treatments for CRC. This integrative approach not only enhances our understanding of CRC biology but also paves the way for personalized medicine, where treatments are tailored to the specific genetic makeup of individual tumors.
In summary, the combination of Expression Atlas data, the critical role of SLC4A4 in CRC, and the strategy of drug repurposing presents a promising avenue for the development of targeted therapies. By focusing on the molecular intricacies of CRC and leveraging existing pharmacological resources, we can advance towards more effective and personalized treatment options for this formidable disease.
2. MATERIALS AND METHODS
2.1. Data Acquisition
To explore the expression profile of the SLC4A4 gene in colorectal cancer (CRC), we performed a comprehensive analysis using the publicly available Expression Atlas database hosted by EMBL-EBI (https://www.ebi.ac.uk/gxa/home). The gene-specific data were retrieved by querying either the gene symbol (SLC4A4) or the Ensembl gene ID (ENSG00000080493). From the gene's overview page, we accessed the “Differential” expression tab, which summarizes gene expression changes across various experimental conditions and tissue types. This tabulated output includes essential metrics such as experiment accession IDs (e.g., E-GEOD series), disease versus control comparisons, log2 fold change (log2FC), and statistically adjusted p-values (adj. p-values) [15].
Subsequently, the results were further scrutinized to identify fold changes in gene expression. This involved comparing the expression levels of SLC4A4 in CRC tissues to those in normal tissues. The analysis was conducted to ensure robust identification of differential expression patterns, facilitating the understanding of SLC4A4's role in CRC.
2.2. Protein Preparation
NBCe1, a sodium-coupled acid-base transporter known as human SLC4A4, was selected for the present study based on several colorectal cancer-related gene expression data collected from the Expression Atlas database. To refine the CryoEM structure of the human SLC4A4 protein (PDB ID: 6CAA) [30] and evaluate it for any structural gaps or missing amino acid residues, homology modeling was carried out using the SWISS-MODEL server [31, 32].
In this approach, the SWISS-MODEL template library (SMTL 2024-03-27 version) was utilized to search for evolutionarily related protein structures that could serve as templates. The most appropriate template was then chosen based on a heuristic selection strategy.
2.3. Screening (Receptor-based)
In the receptor-based screening phase, the target protein's 3D structure (PDB ID: 6CAA), obtained through SWISS-MODEL, was analyzed to identify potential binding pockets. We utilized the DrugRep platform, which is an automated and parameter-free virtual screening server, to facilitate this process. DrugRep employs its in-house, curvature-based cavity detection approach, CurPocket, to automatically predict possible binding pockets on the protein surface. This approach is highly accurate in determining docking boxes for molecular docking. A high-throughput virtual screening was then conducted using a curated library of 2470 FDA-approved drugs sourced from version 5.1.7 of the DrugBank database. The docking workflow was facilitated by DrugRep, which performs batch docking over the drug library using the widely used program AutoDock Vina (version 1.1.2) [33]. The platform automatically determines the docking parameters, including the grid center coordinates and sizes of the search space, based on the predicted pockets. The exhaustiveness parameter was set to 8, the default for DrugRep, which balances computational speed and accuracy. Docking results were ranked by Vina binding affinity scores in kcal/mol. Compounds with docking scores lower than –8.0 kcal/mol were considered promising hits for further analysis. This threshold was selected as a stringent cutoff to identify compounds with strong predicted binding affinities, and it is a common practice in virtual screening studies to filter a large library down to a manageable number of lead candidates [34]. The top hits were then passed to a more refined docking step with CB-Dock2 [35].
2.4. Curation and Preparation of FDA-Approved Ligands for Virtual Screening
The DrugRep cultivated approved drug library refers to a collection of FDA-approved drugs that have been modified for curation and made available in virtual screening campaigns or repurposing studies. This library includes current, up-to-date medications that are already FDA-approved for human use, along with detailed pharmacological profiles and safety data. As part of the high-throughput docking workflow, the online tool DrugRep was used with default parameters to prepare ligands from a curated library of FDA-approved drugs obtained from the DrugBank database (version 5.1.7). This deliberate focus on an established library of FDA-approved drugs is a key strength of our drug repurposing approach, as it eliminates the need for extensive safety and pharmacokinetic studies typically required for novel compounds. The compounds were ranked based on their docking scores, with higher scores indicating stronger predicted interactions with the protein’s binding site. The top three candidates with the most favorable scores were selected for further analysis to identify the most suitable drug candidate [34].
2.5. Molecular Docking Using CB-DOCK2
To refine the docking analysis and confirm the binding poses of the top three candidates (DB01396[Digitoxin], DB00762[Irinotecan], and DB04868[nilotinib]), we utilized the CB-Dock2 web server. CB-Dock2 is a blind docking tool that employs a two-pronged approach: a template-based docking procedure and a structure-based blind docking procedure. The server first attempts to find homologous templates with similar protein and ligand structures to guide the docking process. However, for our specific protein and ligands, the CB-Dock2 server reported that it could not find appropriate templates from its database [35].
Consequently, the docking for all three compounds was performed using the tool's structure-based blind docking pipeline. This procedure, inherited from the original CB-Dock server, operates based on the AutoDock Vina engine and identifies potential binding cavities by analyzing the protein's surface curvature. It then performs molecular docking within these detected cavities to optimize ligand conformations and calculate binding affinities. For each ligand, the server automatically generated multiple binding poses within several predicted cavities. The final results were ranked based on the Vina binding affinity scores (in kcal/mol), and the top-scoring pose for each ligand was selected for further analysis. This approach provided detailed insights into the most probable binding sites and the nature of interactions formed between the ligands and the SLC4A4 protein [35].
2.6. Molecular Dynamics Simulation
Molecular Dynamics (MD) simulations were carried out using GROMACS 2020.1-Ubuntu-2020.1-1 and 2024.1-dev versions (www.gromacs.org) in conjunction with the force field named CHARMM36-jul2022.ff [36]. The initial configuration of the protein-ligand complex was obtained through CB-DOCK2. Preparation of the protein and ligand structures involved the use of the pdb2gmx tool, during which any missing hydrogen atoms were added. Ligand parameters and topology files were generated using the most recent version of CGenFF via the CHARMM-GUI interface [37]. The simulation system was solvated in a cubic water box using the TIP3P water model, maintaining a minimum distance of 10 Å between the protein and the box boundaries. Sodium ions were introduced to achieve charge neutrality.
Periodic boundary conditions were applied to both systems using the Leapfrog integrator over a 50-nanosecond run, incorporating both NVT and NPT equilibration phases. Electrostatic interactions were computed using the Particle-Mesh Ewald (PME) method. A force-based switching function was applied to truncate non-bonded interactions at distances greater than 10 and 12 Å. Energy minimization was performed using the steepest descent algorithm for 50,000 steps to eliminate any steric clashes or unfavorable contacts. Structural analyses, including Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuations (RMSF), were conducted using the gmxrms and gmxrmsf utilities. Hydrogen bond analysis was performed using the gmxhbond tool. Additional metrics, such as the radius of gyration (Rg) and Solvent-Accessible Surface Area (SASA), were determined using the gmx gyrate and gmx sasa commands, respectively. Visualization of the simulation data was performed in PyMOL, while graphical representations were generated using Grace software [38, 39]. The ligand-receptor dynamics were simulated for 50 ns on the GridMarkets platform, taking approximately 22 hours.
2.7. Study Design
This study was designed as an exploratory, quantitative, and computational investigation that integrates public transcriptomic data with in silico drug discovery tools. The primary objective was to assess the role of the SLC4A4 gene in colorectal cancer (CRC) and to identify potential FDA-approved drugs that could target its protein product.
The study followed a structured multi-phase computational pipeline:
- Gene Expression Analysis – Public datasets were mined from the Expression Atlas database to determine differential expression patterns of SLC4A4 in CRC vs. normal tissue using log2 fold change and adjusted p-values.
- Protein Modeling – The Cryo-EM structure of the SLC4A4 protein (PDB ID: 6CAA) was refined using SWISS-MODEL.
- Virtual Screening and Docking – A library of FDA-approved drugs from DrugBank (via DrugRep) was docked against the modeled protein using AutoDock Vina, and redocked with CB-DOCK2.
- Molecular Dynamics Simulation – The top drug candidate (Nilotinib) was subjected to a 50-nanosecond MD simulation using GROMACS to validate binding stability through RMSD, RMSF, hydrogen bonding, radius of gyration, and SASA metrics.
The study is observational in nature, with no live subjects, and all data were sourced from publicly available databases. All tools used are open-source or web-based. The methodology ensures reproducibility and transparency in computational drug repurposing studies.
3. RESULTS
3.1. Expression of SLC4A4 from Expression Atlas Datasets
To explore the expression of the SLC4A4 gene in colorectal cancer (CRC), we conducted an extensive analysis using RNA sequencing (RNA-seq) or array-based data from the Expression Atlas database. Upon refining the analysis to focus specifically on CRC datasets, we identified significant differential expression patterns of SLC4A4. The results consistently indicated a downregulation of SLC4A4 in colorectal cancer samples compared to normal tissues. This was determined using a Log2 Fold Change (log2FC) threshold of less than -1 and an adjusted p-value (adj. p-value) of less than or equal to 0.05. This downregulation was observed across multiple CRC datasets and depicted in Table 1, highlighting a potential role of SLC4A4 in CRC pathogenesis.
3.2. Molecular Modelling
A 3D model of SLC4A4, with reference to the template Q9Y6R1, underwent modeling as described in Table 2. The Protein Structure and Model Assessment Tool of the Swiss-Model Server was used to evaluate the structure, yielding favorable scores: Global Model Quality Estimate = 0.71; Ramachandran Favoured (%) = 90.69. In addition, the MolProbity Score was 1.64 (Table 3). These assessments emphasized the potential of using this model for subsequent structure-based 6CAA analysis and improvement.
| Gene | Species | Experiment accession | Comparison | log_2 fold change | Adjusted p-value |
|---|---|---|---|---|---|
| ENSG00000080493 | homo sapiens | E-GEOD-19249 | 'colon cancer' vs 'normal' in 'colon; Fresh-frozen tissue' | -5.9 | 2.18433988997483E-05 |
| ENSG00000080493 | homo sapiens | E-GEOD-76987 | 'colon adenocarcinoma' vs 'normal' | -4.4 | 5.53014902194911E-40 |
| ENSG00000080493 | homo sapiens | E-GEOD-68086 | 'colorectal carcinoma' vs 'normal' | -3.2 | 2.94812359574088E-09 |
| ENSG00000080493 | homo sapiens | E-GEOD-19249 | 'colon cancer' vs 'normal' in 'colon; Formalin-fixed paraffin-embedded tissue' | -3.1 | 0.02006192248942 |
| ENSG00000080493 | homo sapiens | E-GEOD-4183 | 'colon adenoma' vs 'none' | -3 | 6.23893701946427E-05 |
| ENSG00000080493 | homo sapiens | E-GEOD-50760 | 'colorectal cancer metastatic in the liver' vs 'normal' | -2.5 | 2.04883548287025E-06 |
| ENSG00000080493 | homo sapiens | E-GEOD-4183 | 'colorectal cancer' vs 'none' | -2.2 | 0.049104604966632 |
| Template | Seq Identity | Oligo-state | Found by | Method | Resolution | Seq Similarity | Range | Coverage | Description |
|---|---|---|---|---|---|---|---|---|---|
| Q9Y6R1.1.A | 99.5 | monomer | AFDB search | AlphaFold v2 | --- | 0.61 | 35 - 1035 | 0.97 | Electrogenic sodium bicarbonate cotransporter 1 |
| Model | Built with | Oligo-State | Ligands | GMQE | MolProbity Score | Ramachandran Favoured |
|---|---|---|---|---|---|---|
| 1 | ProMod3 3.4.0 | monomer | None | 0.71 | 1.64 | 90.69% |
3.3. Molecular Docking
Virtual screening of approved drugs was performed using the DrugRep tool with default settings, utilizing compounds from the DrugBank database (version 5.1.7). The virtual screening and docking results revealed a key binding pocket on the SLC4A4 protein, which was a primary target for ligand binding. The characteristics of this pocket are detailed in Table 4. The pocket, identified as Pocket 1, has a significant volume of 3676 Å3 and is located at the coordinates -1.4, -18.4, -0.4. It is composed of key amino acid residues, including both hydrophilic residues like Aspartic Acid (ASP), Glutamic Acid (GLU), and Lysine (LYS), and hydrophobic residues such as Phenylalanine (PHE) and Leucine (LEU). This diverse composition suggests that the pocket can accommodate a variety of ligands and form multiple types of interactions, which is consistent with the favorable binding affinities observed for our top drug candidates.
To derive the top-ranked docking pose according to scoring metrics, redocking was performed using CB-DOCK2 with the selected compounds: DB01396 (Digitoxin), DB00762 (Irinotecan), and DB04868 (Nilotinib). Notably, Nilotinib exhibited the highest docking score, suggesting a strong binding affinity (Table 6).
| Volume | Center | Size | |
|---|---|---|---|
| 3676 | -1.4,-18.4,-0.4 | 22,21,19 | Chain A (ASP960 ASN763 ASP966 VAL760 VAL961 ASP947 GLY387 PHE531 LYS974 PHE536 VAL533 PRO963 GLN952 LEU392 GLU971 LYS970 HSD386 LEU955 ALA773 ASP959 LYS765 GLU766 SER956 LYS768 LYS944 TYR775 ARG764 LEU769 GLU541 CYS389 ILE962 ARG943 GLN534 ARG538 GLY772 LYS967 TYR535 GLU964 GLU391 VAL940 THR537 ASP388) |
| DrugBank ID | Docking Score (kcal/mol) |
|---|---|
| DB01396 | -10.2 |
| DB00762 | -10 |
| DB04868 | -9.6 |
| FDA-approved drugs | Pocket ID |
Vina score (kcal/mol) |
Cavity volume (Å3) |
Center (x, y, z) |
Docking size (x, y, z) |
Pocket Seq |
|---|---|---|---|---|---|---|
| DB04868 (Nilotinib) | C1 | -11.2 | 3676 | -1, -18, 0 | 24, 24, 24 | Chain A: HIS386 GLY387 ASP388 CYS389 GLU391 LEU392 GLN534 PHE536 THR537 ARG538 GLU541 VAL760 ASN763 ARG764 LYS765 GLU766 LYS768 TYR775 GLN877 PHE895 ILE896 TYR897 HIS900 ARG943 LYS944 GLN952 HIS953 LEU955 SER956 PHE957 ASP959 ASP960 VAL961 ILE962 PRO963 GLU964 LYS965 LYS967 LYS968 GLU971 |
| DB00762 (Irinotecan) | C3 | -10 | 2519 | -9, 4, -1 | 30, 30, 30 | Chain A: LYS233 ASN422 ILE423 GLN424 SER427 LYS670 SER672 PRO673 PHE675 THR677 THR678 ARG680 LYS681 LEU682 ASP685 PHE686 GLU766 HIS767 ASP809 LYS812 MET813 GLU814 THR815 GLU816 THR817 SER818 ALA819 PRO820 GLY821 GLU822 GLN823 PRO824 PHE826 ARG830 GLY875 |
| DB01396 (Digitoxin) | C4 | -9.8 | 2046 | -23, 2, 27 | 33, 33, 33 | Chain A: TRP87 ILE88 LYS89 PHE90 LYS93 VAL94 GLN96 SER102 HIS105 VAL106 ALA107 THR108 LEU109 SER110 LEU111 LYS291 HIS294 GLU295 ARG298 THR302 SER305 ASP306 GLU307 GLU328 PHE329 GLU332 VAL333 ILE334 VAL335 LEU336 PRO338 TRP341 ASP342 PRO343 ALA344 ARG346 ILE347 GLU348 PRO349 PRO350 LYS351 SER352 LEU353 PRO354 SER355 SER356 ASP357 LYS358 ARG359 |
Protein 6CAA and Nilotinib docking complex.
The molecular docking complex formed between the modeled protein 6CAA and Nilotinib (Fig. 1) was subsequently subjected to molecular dynamics simulation to evaluate its stability and interaction profile.
3.4. Molecular Dynamics Simulation
In the case of the protein–nilotinib complex, the system exhibited stable dynamics, with the protein displaying a maximum RMSD of approximately 1.5 nanometers over the 50-ns simulation (represented by the black line in Fig. (2A). The ligand maintained fluctuations within ~0.30 nm RMSD throughout the same period (red line in Fig. 2A), indicating consistent positioning within the binding pocket. Overall, the RMSD analysis indicates a stable interaction between the ligand and the protein's active site.
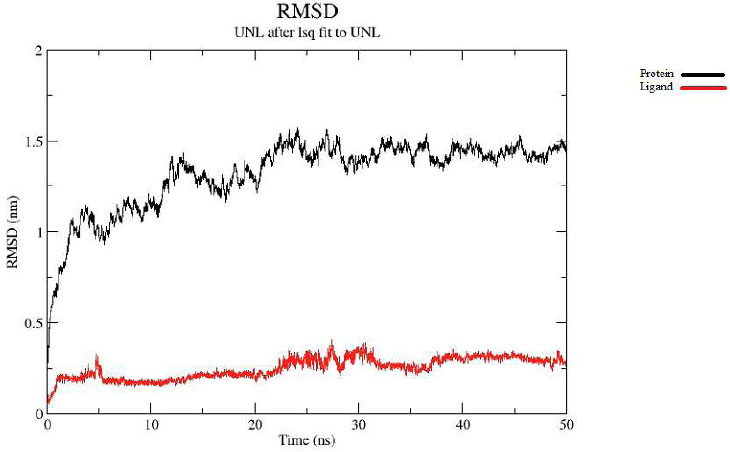
Protein-ligand RMSD.
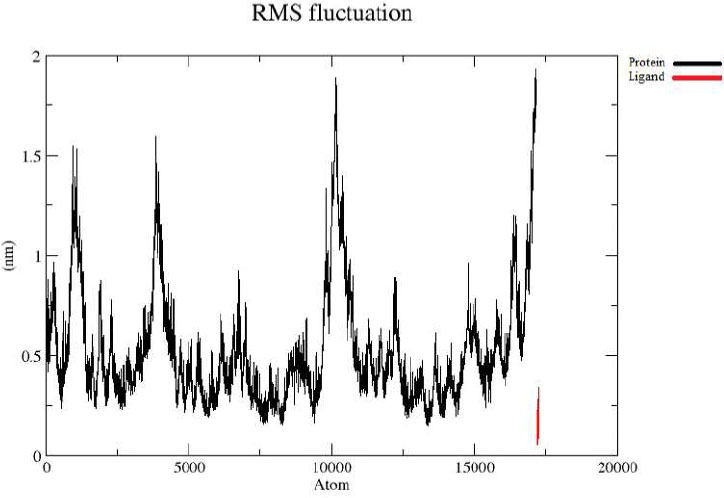
Protein-ligand RMSF.
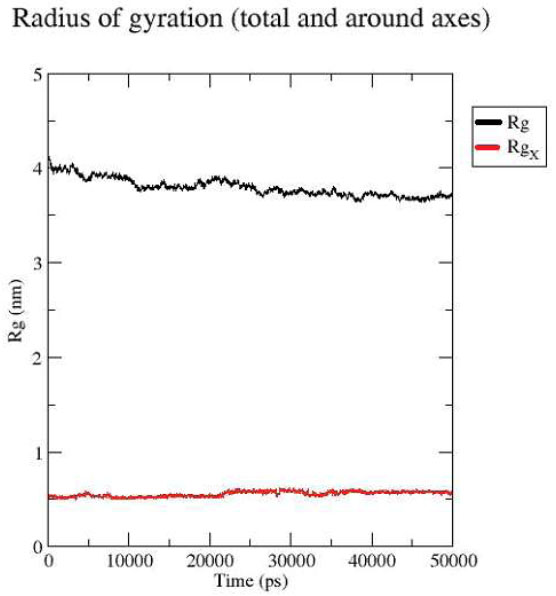
Protein-ligand Radius of Gyration.
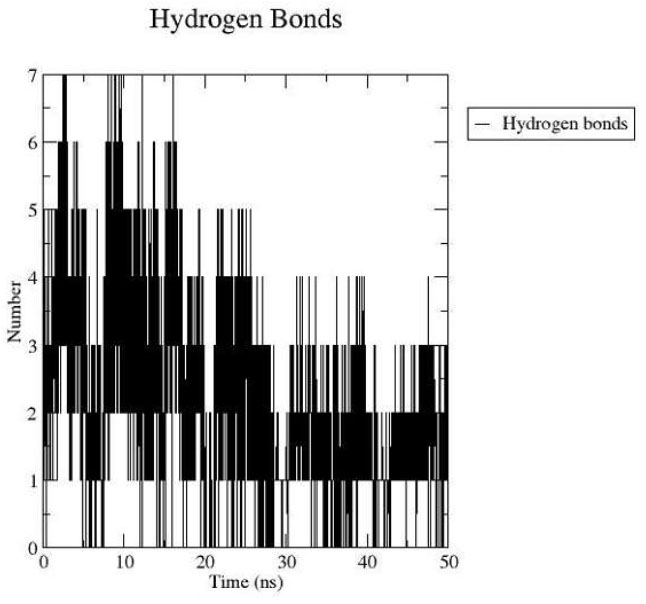
Protein-ligand Hydrogen bond interaction.
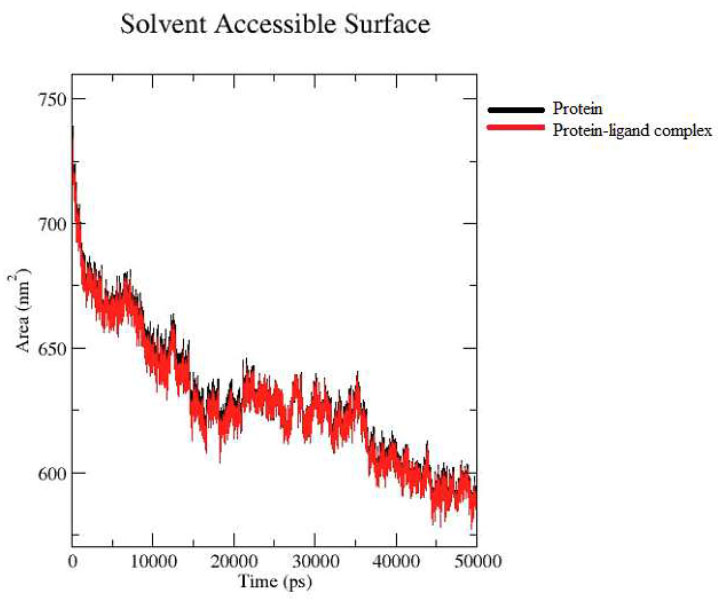
Protein-ligand SASA analysis.
The RMSF analysis revealed that the N-terminal region of the protein displayed minor fluctuations around 1.0 nm, while the C-terminal showed increased flexibility with fluctuations reaching up to ~2.0 nm, as shown by the black line in Fig. (2B). In contrast, the ligand exhibited relatively low flexibility, remaining within ~0.5 nm throughout the simulation, indicated by the red line.
The radius of gyration (Rg) analysis revealed that the ligand, represented by the red line, fluctuated up to 0.5 nm, whereas the protein, indicated by the black line, exhibited a gradual decrease from 4.0 nm to approximately 3.8 nm by the end of the simulation. These results suggest that the ligand is positioned stably within the protein's binding region Fig. (2C).
Hydrogen bond analysis revealed that the ligand formed a maximum of seven hydrogen bonds at various points during the simulation, specifically at 4, 8, 9, 11, 12, 14, and 16 nanoseconds. By the end of the 50-nanosecond simulation, three hydrogen bonds remained within the protein-ligand complex, as illustrated in Fig. (2D).
The Solvent-Accessible Surface Area (SASA) analysis showed the protein alone (black line) and the protein-ligand complex (red line) throughout the duration of the MD simulation. The close overlap of the two graphs indicates minimal fluctuations, suggesting that the system remained stable throughout the entire simulation period Fig. (2E).
Following energy minimization, the Nilotinib–protein complex exhibited several key interactions. At the initial conformation (0 ns), the compound formed hydrogen bonds with Ser38, Arg40, Pro105, Ile132, Gln107, Ser349, and Glu351. Pi-interactions were observed with residues such as Arg40, Ala104, Lys277, Pro279, His351, Thr861, Ser862, Ala863, and Lys869. Additionally, van der Waals interactions involved Val35, Lys37, Tyr39, Arg41, Pro106, Leu108, Lys133, Met348, Thr859, and Glu861 Fig. (3A).
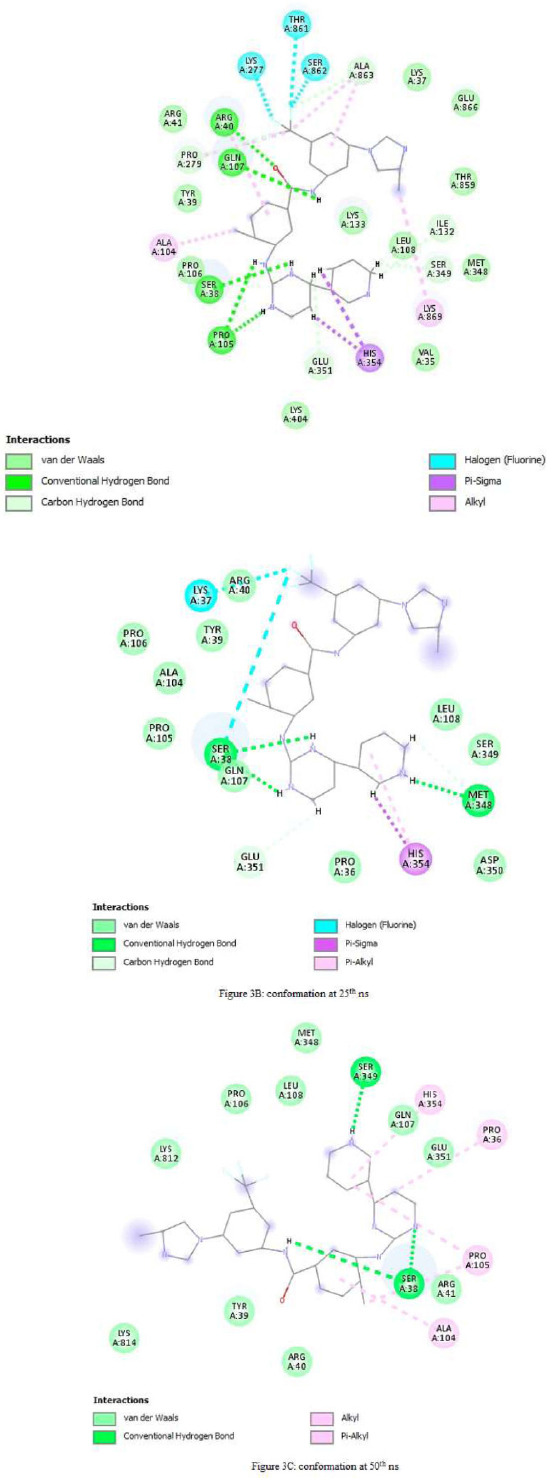
At the 25 ns time point, Nilotinib maintained four hydrogen bonds with Cys38 and Glu351. Pi-interactions were noted with Lys37, Ser38, and His354, while van der Waals interactions occurred with Pro36, Tyr39, Arg40, Ala104, Pro105, Pro106, Gln107, Leu108, Ser349, and Asp350 (Fig. 3B).
By the end of the simulation at 50 ns, the ligand formed hydrogen bonds with Ser38 and Ser349. Pi-interactions were identified with Pro36, Ala104, Pro105, and His354, and van der Waals forces were established with Tyr39, Arg40, Arg41, Pro106, Gln107, Leu108, Glu351, and Lys814 (Fig. 3C).
Across all time points—0 ns, 25 ns, and 50 ns—Nilotinib consistently remained within the binding pocket of the protein, indicating stable occupancy and a range of favorable interactions throughout the simulation.
4. DISCUSSION
While the role of SLC4A4 in maintaining cellular pH balance is well-established, its biological significance in colorectal cancer extends far beyond this fundamental function. Recent studies have highlighted its crucial involvement in modulating the tumor microenvironment and influencing key processes such as Epithelial-Mesenchymal Transition (EMT) and metastasis.
Beyond its direct effects on cancer cells, SLC4A4 plays a critical role in shaping the inflammatory TME. Studies have shown that SLC4A4 expression positively correlates with the infiltration of various immune cells, including CD8+ T cells, dendritic cells, macrophages, and NK cells, in colorectal cancer (CRC) tissues [18, 31]. This is significant because these immune cells are crucial for an effective anti-tumor immune response. Downregulation of SLC4A4, therefore, can lead to a less inflammatory, more immunosuppressive TME, which allows cancer cells to evade immune surveillance and thrive. This finding suggests that restoring or maintaining SLC4A4 function could be a novel strategy to “re-educate” the TME, making it more hostile to cancer cells and more receptive to immunotherapy. Furthermore, research has indicated that higher SLC4A4 expression may promote a response to certain immunotherapies, such as Nivolumab and Ipilimumab, while its downregulation may favor a response to other targeted therapies [31]. This dual function highlights SLC4A4's potential not only as a therapeutic target itself but also as a predictive biomarker to guide treatment selection and personalize patient care.
The biological justification for targeting SLC4A4 is further reinforced by its involvement in the Epithelial-Mesenchymal Transition (EMT) process, a critical step in cancer metastasis. EMT is a complex cellular program where epithelial cells lose their cell-to-cell adhesion and acquire migratory and invasive properties, enabling them to break away from the primary tumor and spread to distant sites. Research indicates that low SLC4A4 expression correlates with increased lymph node and distant metastasis and regulates partial EMT phenotypes that are essential for cancer cell migration and invasion. This suggests that SLC4A4 acts as a tumor suppressor by inhibiting EMT. By targeting and restoring SLC4A4's function, it may be possible to block this key metastatic pathway, thereby preventing the spread of CRC and improving patient outcomes [17]. This adds a powerful layer to the rationale for targeting SLC4A4, as it addresses one of the most lethal aspects of cancer progression.
The downregulation of SLC4A4 observed in colorectal cancer (CRC) samples compared to normal tissues suggests that this gene may play a crucial role in the pathogenesis of CRC [40]. SLC4A4 encodes a sodium bicarbonate cotransporter involved in pH regulation and cellular homeostasis [41]. Its altered expression has been implicated in various cancers, including CRC, highlighting its potential as a therapeutic target [42, 43].
Drug repurposing, the strategy of identifying new uses for existing drugs, presents an efficient and cost-effective approach to cancer treatment. In this context, targeting SLC4A4 could offer new therapeutic avenues. The identification of specific inhibitors or modulators of SLC4A4 could help restore normal cellular functions and inhibit cancer progression [19]. Moreover, targeting SLC4A4 could potentially disrupt the tumor microenvironment, making cancer cells more susceptible to existing treatments.
Nilotinib is a second-generation tyrosine kinase inhibitor that was initially developed and approved for the treatment of chronic myeloid leukemia due to its high specificity and efficacy against the BCR-ABL fusion protein. However, accumulating evidence from preclinical studies suggests that nilotinib possesses broader anti-cancer properties, making it a promising candidate for repurposing in other malignancies, including colorectal cancer (CRC) [44].
Mechanistically, nilotinib has demonstrated the ability to inhibit several tyrosine kinases implicated in cancer progression. Notably, recent research has identified Discoidin Domain Receptor 1 (DDR1) as a key target of nilotinib in CRC. DDR1 is involved in mediating tumor cell invasion and metastasis by activating downstream pathways, such as β-catenin signaling. In vitro and in vivo studies have shown that nilotinib effectively suppresses CRC cell invasion and reduces metastatic burden by blocking DDR1 activity and its downstream effects [45, 46].
Beyond its direct anti-tumor effects, nilotinib also exhibits immunomodulatory properties that may enhance its therapeutic potential in CRC [18]. Dong et al. (2024) demonstrated that nilotinib can restore the expression of Major Histocompatibility Complex class I (MHC-I) molecules in CRC cells, thereby enhancing tumor immunogenicity and improving the response to immune checkpoint inhibitors, such as anti-PD-L1 therapy. This dual mechanism—direct inhibition of tumor-promoting kinases and enhancement of anti-tumor immunity positions nilotinib as a particularly attractive agent for combination therapy strategies in CRC [47].
Importantly, nilotinib’s clinical safety and pharmacokinetic profiles are well established from its extensive use in chronic myeloid leukemia patients. Long-term clinical studies have demonstrated that nilotinib is generally well-tolerated, with manageable adverse effects and a predictable pharmacological profile [48]. These attributes support its potential for rapid clinical translation and repurposing in CRC, as dosing regimens and safety monitoring protocols are already well defined [49].
In summary, nilotinib’s demonstrated efficacy in targeting DDR1-mediated signaling, its ability to enhance anti-tumor immunity, and its established safety profile make it a compelling candidate for further investigation as a therapeutic agent in colorectal cancer. To our knowledge, this is the first study to apply an in silico drug repurposing strategy using FDA-approved drugs to identify potential inhibitors of SLC4A4 in colorectal cancer. While SLC4A4 has been studied in relation to pH regulation and prognosis, no previous work has evaluated FDA-approved drugs for direct targeting of this transporter, which positions this study as a novel exploratory step in CRC drug discovery.
5. CONCLUSION
In conclusion, the docking and molecular dynamics simulations of nilotinib with the modeled 6CAA protein have yielded several significant findings, including high affinity and stability, consistent binding poses, protein flexibility, strong hydrogen bonding, and stable conformation, as mentioned in the results section.
The conformational analysis of the MD simulation results confirmed the presence of nilotinib within the binding site region of the protein. Considering these findings, nilotinib emerges as a promising candidate for further investigation and potential development due to its high affinity for binding strongly and stably without significantly distorting the protein conformation.
Further research is necessary to validate the effectiveness of nilotinib in colorectal cancer (CRC) and to elucidate the precise mechanisms by which it interacts with SLC4A4 and other relevant molecular targets. Nevertheless, the repurposing of nilotinib for CRC treatment holds significant promise and could lead to more effective and personalized therapeutic strategies.
Limitations
Despite these encouraging results, several limitations must be considered. First, this study is entirely computational and lacks experimental validation. As such, its findings should be regarded as a foundation for future research. The predictions may be influenced by the potential for off-target effects and the inherent limitations of current protein modeling and docking protocols, which may not accurately capture the dynamic complexity of protein-ligand interactions within a cellular context. Additionally, we acknowledge the inability to include external control ligands or known inhibitors for benchmarking, as the DrugRep server used in this study only allows screening of its curated FDA-approved drug library and does not permit user-defined ligand input. Furthermore, no benchmarking against a known SLC4A4 inhibitor was possible, as no such inhibitors are currently reported. Likewise, binding free energy calculations (MM-PBSA or MM-GBSA) were not performed, which would provide additional energetic insights. These will be pursued in future studies alongside experimental bioassays. Future studies using customizable docking platforms could address this by incorporating specific controls for enhanced benchmarking. Overall, future work should include experimental validation and exploration of combination strategies to fully realize the therapeutic potential of nilotinib and other candidates targeting SLC4A4 in CRC.
AUTHORS’ CONTRIBUTIONS:
K.P., P.P.G., R.R.K.N.: Conceptualization, methodology, formal analysis, investigation, resources, data curation, Writing, draft preparation; S.L.K., P.S.S., R.R.K.N.: Supervision;.
LIST OF ABBREVIATIONS
| CRC | = Colorectal Cancer |
| SLC4A4 | = Solute Carrier Family 4 Member 4 |
| pH | = Potential of Hydrogen (measure of acidity/basicity) |
| p-EMT | = Partial Epithelial–Mesenchymal Transition |
| MHC-I | = Major Histocompatibility Complex Class I |
| PD-L1 | = Programmed Death-Ligand 1 |
| MD | = Molecular Dynamics |
| RMSD | = Root Mean Square Deviation |
| RMSF | = Root Mean Square Fluctuation |
| Rg | = Radius of Gyration |
| SASA | = Solvent Accessible Surface Area |
| PME | = Particle Mesh Ewald (method for computing long-range electrostatics) |
| TIP3P | = Transferable Intermolecular Potential with 3 Points (water model) |
| NVT | = Constant Number of Particles, Volume, and Temperature |
| NPT | = Constant Number of Particles, Pressure, and Temperature |
AVAILABILITY OF DATA AND MATERIALS
The data and supportive information are available within the article.
ACKNOWLEDGEMENTS
Declared none.


