Molecular Dynamics Studies of the Bufallo Prion Protein Structured Region at Higher Temperatures
Abstract
Background:
Molecular Dynamics (MD) studies of Buffalo Prion Protein (BufPrPC) (J Biomol Struct Dyn 2016; 34(4): 762-77) showed that the structure of this protein is very stable at room temperature (whether under neutral pH or low pH environments).
Methods:
In order to understand the reason why buffalo is resistant to prion diseases and why BufPrPC is so stable at room temperature, this paper will prolong our MD running time at room temperature and extend our research to higher temperatures to study this BufPrPC structure furthermore.
Results:
From the salt bridge point of view, we found an important reason why BufPrPC is so stable at room temperature; this might be a nice clue of drug discovery or drug design for the treatment of prion diseases.
Conclusion:
In conclusion, this brief article talks about the MD results of BufPrP at different temperatures and presents a clue to seek the reasons for the conversion from normal cellular prion protein (PrPC) to diseased infectious prions (PrPSc). This should be very useful for the goals of medicinal chemistry in prion diseases research fields.
1. INTRODUCTION
Unlike bacteria and viruses, which are based on DNA and RNA, prions are unique as disease-causing agents since they are misfolded proteins. Prions propagate by deforming harmless, correctly folded proteins into copies of themselves. The misfolding is irreversible. Prions attack the nervous system of the organism, causing an incurable, fatal deterioration of the brain and nervous system until death occurs. Some examples of these diseases are mad cow disease in cattle, chronic wasting disease in deer and elk, and Creutzfeldt-Jakob disease in humans.
Not every species is affected by prion diseases. Water buffalo is a species being resistant to prion diseases. The research question arises from the molecular structure point of view as to what is the reason that allows it to retain its molecular structure folding? This is the research question addressed in this paper. Many experimental studies have shown that BufPrP is very stable so that it resists the infection of diseased prions [1-6]. The brief review of these experimental studies has been done earlier [7]. In addition, in 2015, Zhao et al. [8] reported that the prion protein gene polymorphisms associated with bovine spongiform encephalopathy susceptibility differ significantly between cattle and buffalo [8] and reported three significant findings in buffalo: 1) extraordinarily low deletion allele frequencies of the 23- and 12-bp indel polymorphisms; 2) significantly low allelic frequencies of six octarepeats in coding sequence; and 3) the presence of S4R, A16V, P54S, G108S, V123M, S154N and F257L substitutions in buffalo coding sequences [8]. In 2017, Zhao et al. [9] reported that fixed differences in the 3'UTR of buffalo PRNP gene provide binding sites for miRNAs post-transcriptional regulation [9]. In 2017, Yaman et al. [10] investigated the prion protein gene (PRNP) polymorphisms in Anatolian, Murrah, and crossbred water buffaloes (Bubalus bubalis).Yaman et al. [10, 11] reported the T/A and T/G genotypes in water buffaloes for the first time; they found: 1) three synonymous single nucleotide polymorphisms (SNP) at positions 126, 234, and 285, and a non-synonymous SNP at position 322 (G108S); 2) triplet G/A/T base substitutions at position 126 and two additional genotypes, T/A and T/G, also at this position; and 3) six octarepeats that indicated the presence of the wild-type PRNP6 allele in the coding region [11].
As we all know, prion diseases are caused by the conversion from normal cellular prion protein (PrPC) to diseased infectious prions (PrPSc); in structure, the conversion is mainly from α-helices to β-sheets (generally, PrPC has 42% α-helix and 3% β-sheet, but PrPSc has 30% α-helix and 43% β-sheet) [12-17]. The structural region of a PrPC usually consists of β-strand 1 (β1), α-helix 1 (H1), β-strand 2 (β2), α-helix 2 (H2), α-helix 3 (H3), and the loops link them to each other. The conformational changes may be amenable to study by molecular dynamics (MD) techniques. NMR experiences showed that a prion resistant species does not have higher conformational stability at a higher temperature than non-resistant species [18]; this means that at high temperature, the α-helices of PrPC will turn to β-sheets of PrPSc so that we can find out some secrets of the protein structural conformational changes of PrP. Hence, in this paper, we will use MD to study the molecular structure of buffalo prion protein Buf PrPC(124-227) [7]. In a research study [7], the structure of BufPrPC(124-227) has been shown to be very stable at room temperature, whether under neutral pH or low pH environments. In order to understand the reason why buffalo is resistant to prion diseases and why BufPrPC is so stable at room temperature, this paper will prolong our MD running time at room temperature and extend our research to higher temperatures to study this BufPrPC structure, as presented in the Methods and Materials section. In the section of Results and Discussion, we will analyze our MD computational results and discuss the reason why BufPrPC is so stable at room temperature. The Conclusions section presents a concluding remark of this paper and proposes a nice clue from BufPrP studies for drug discovery/design of the treatment of prion diseases.
2. MATERIALS AND METHODS
The MD structure used for the paper is the region BufPrP structure, which has also been used earlier [7]. Earlier [7], 25 ns' MD simulations were done at room temperature 300 K. This paper prolongs the MD running time to 30 ns. Moreover, this paper extends 30 ns' MD simulations to higher temperatures of 350 K and 450 K, respectively.
The MD methods for 350 K [19] and 450 K [20-24] are completely the same [20-25]. For the neutral pH environment, we let CYS, HIS change into CYX, HID respectively and add Na+ ions; and we change CYS, HIS, ASP, GLU into CYX, HIP, ASH, GLH, respectively, in the low pH environment and add Cl- ions and NME. We emphasize that all our methods are completely reproducible [26].
BufPrP has stable molecular structures at 300 K during the whole 25 ns' MD simulations [7], where the Root Mean Square Deviation (RMSD) and Radius of Gyration values are not changing very much during the whole 25 ns. As we all know, RMSD and Radius of Gyration are two indicators for structural changes in a protein. The Radius of Gyration is the mass-weighted scalar length of each atom from the Center-of-Mass (COM). The RMSD is used to measure the scalar distance between atoms of the same type for two structures. In this paper, the initial structures compared with all the MD structures are the minimized/optimized structures. From the RMSD and Radius Of Gyration observations for 300 K, 350 K and 450 K, we then carry out deeper researches on the secondary structures developments during the whole 30 ns' MD simulations of 300 K, 350 K, 450 K. The BufPrP molecular structure is maintained by a network of atoms by their peptide bonds, covalent bonds (e.g., the disulfide bond S-S between Cys179 and Cys214), and noncovalent bonds such as hydrogen bonds, salt links, van der Waals contacts, and hydrophobic interactions. We will find out which bonds contribute to maintain the stability of BufPrP.
3. RESULTS AND DISCUSSION
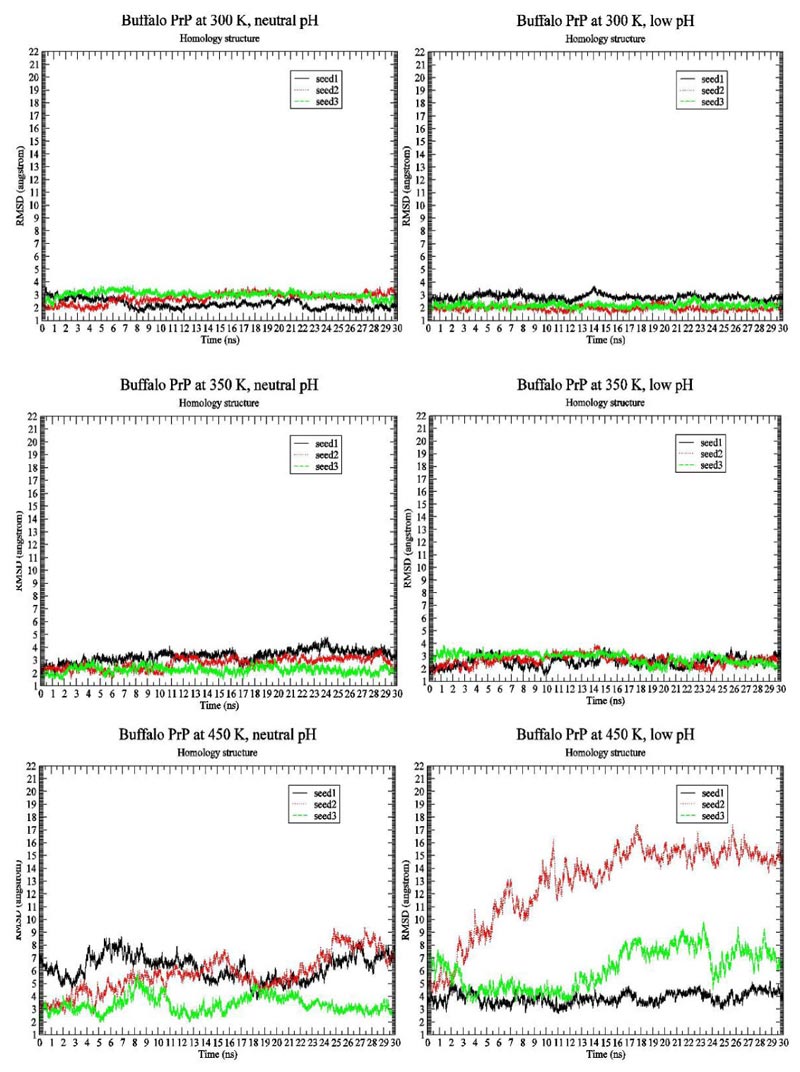
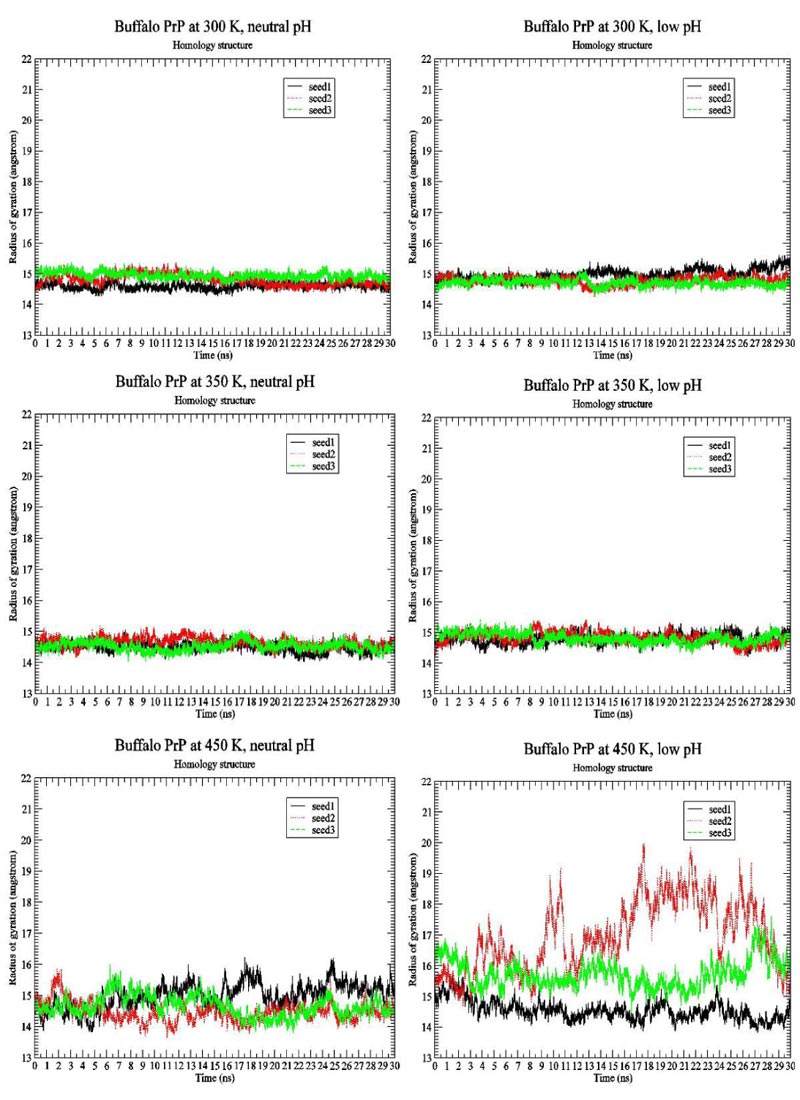
We first present the RMSD Fig. (1) and Radius of Gyration Fig. (2) results of BufPrP at 300 K, 350 K and 450 K, respectively, during the whole 30 ns.
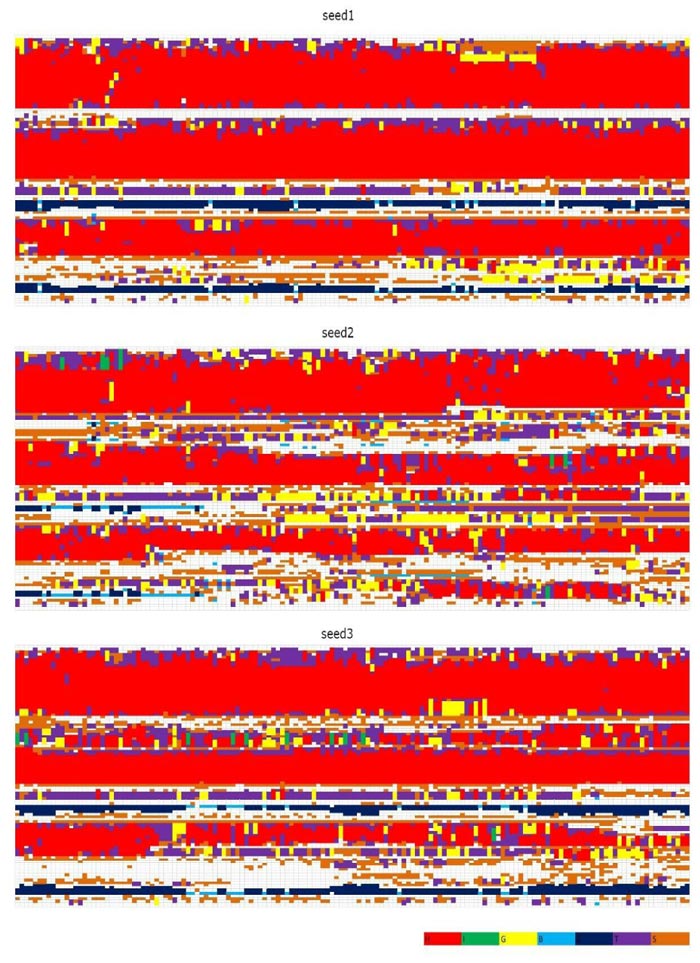
In Figs. (1 and 2) [27], we see that (i) at 300 K and 350 K, the values of RMSD and Radius of Gyration are not changing very much (RMSDs varying within 2 angstroms and Radius of Gyrations varying within 1 angstrom ; these are within normal variations of MD structures because typically we would want our RMSD to be less than 1.5-2 angstroms), whether under neutral or low pH environments, (ii) the RMSD performance occurs at 450 K under neutral pH environment but RMSDs vary largely within 7 angstroms (however from Fig. (3), we think the α-helices are still unfolded under neutral pH environment at 450 K because variations of Radius Of Gyrations are normally within 2 angstroms), and the Radius Of Gyration performance of 450 K under neutral pH environment is slightly worse than at 300 K and 350 K but still normally varying within 2 angstroms; and (iii) at 450 K under low pH environment, for seed2, it is clear that the α-helices are unfolded into β-structures. Thus, next, we just see the Secondary Structure graphs of BufPrP at 450 K, with a low pH value Fig. (3). The Secondary Structures here are H - the α-helix, I - the 5 helix called π-helix, G - the 3-helix called 310-helix, B - the residue in isolated β-bridge, E - the extended strand (participates in β-ladder), T - the hydrogen bonded turn, and S - the bend. The colours and the developments of H, I, G, B, E, T and S are illuminated in Fig. (3).
Seeing Fig. (3), we know that not only for seed 2 but also for seed 3, α-helices H1 and H2 unfold into other forms of secondary structures. In summary, below, we may only focus on a low pH environment at 450 K to find out the reasons for the unfolding of α-helices H1 and H2.
Because the change of pH environments from neutral pH to low pH will lead to the loss of salt bridges, we will mainly analyse the noncovalent bonds of Salt Bridges (SBs) as follows (Tables 1-2).
| Buffalo PrP / | - | 300 K | - | - | 350 K | - | - | 450 K | - |
|---|---|---|---|---|---|---|---|---|---|
| Salt Bridges (SBs) | Seed1 | Seed2 | Seed3 | Seed1 | Seed2 | Seed3 | Seed1 | Seed2 | Seed3 |
| ASP147@CG-ARG148@CA.CZ | 100 | 100 | 100 | 100 | 100 | 100 | 99.95 | 99.97 | 99.98 |
| HIS155@CG-ARG156@CA.CZ | 99.74 | 99.80 | 99.63 | 99.80 | 99.87 | 99.75 | 99.72 | 99.40 | 99.50 |
| HIS155@NE2-ARG156@CA.CZ | 5.18 | 6.53 | 7.49 | 31.90 | 46.75 | 13.26 | 24.63 | 18.23 | 22.53 |
| GLU211@CD-ARG208@CA.CZ | 99.47 | 99.70 | 93.46 | 91.26 | 98.07 | 99.67 | 97.08 | 97.38 | 95.88 |
| GLU207@CD-LYS204@CA.NZ | 98.48 | 99.88 | 99.86 | 99.19 | 98.05 | 98.63 | 87.38 | 97.52 | 97.67 |
| GLU221@CD-ARG220@CA.CZ | 96.78 | 63.88 | 52.51 | 96.77 | 87.26 | 94.58 | 69.02 | 69.40 | 77.25 |
| GLU186@CD-LYS185@CA.NZ | 93.63 | 92.88 | 96.30 | 68.63 | 84.71 | 86.76 | 19.93 | 77.72 | 74.98 |
| ASP178@CG-ARG164@CA.CZ | 87.89 | 23.89 | 1.51 | 38.56 | 33.93 | 47.25 | 4.30 | 30.45 | 39.87 |
| GLU196@CD-LYS194@CA.NZ | 70.57 | 60.08 | 19.20 | 28.13 | 20.43 | 5.61 | 4.65 | 26.40 | 25.12 |
| GLU207@CD-RG208@CA.CZ | 57.4 | 35.53 | 73.37 | 51.93 | 66.71 | 62.95 | 81.35 | 78.92 | 77.82 |
| ASP147@CG-IS140@ND1.HD1 | 38.91 | 20.51 | 15.12 | 46.31 | 52.45 | 64.40 | 0.10 | 49.72 | 25.20 |
| GLU152@CD-ARG148@CA.CZ | 34.72 | 23.28 | 30.46 | 50.64 | 38.77 | 40.89 | 46.23 | 19.42 | 28.72 |
| GLU152@CD-ARG151@CA.CZ | 33.62 | 36.68 | 31.59 | 39.79 | 34.66 | 41.93 | 40.73 | 50.43 | 37.97 |
| ASP144@CG-ARG148@CA.CZ | 27.93 | 85.55 | 75.43 | 32.47 | 52.25 | 49.01 | 2.48 | 34.45 | 52.88 |
| ASP147@CG-ARG151@CA.CZ | 19.40 | 48.61 | 27.63 | 27.83 | 20.75 | 25.65 | 2.98 | 51.53 | 51.65 |
| HIS187@NE2-ARG156@CA.CZ | 14.09 | 59.43 | 68.19 | 64.04 | 18.17 | 53.34 | - | 21.77 | 14.53 |
| HIS187@CG-ARG156@CA.CZ | 0.04 | 0.13 | 0.33 | 5.29 | 0.35 | 11.79 | - | 1.03 | 4.23 |
| GLU221@CD-ARG164@CA.CZ | - | 38.33 | 6.32 | 0.02 | 0.25 | 0.33 | 0.48 | 0.33 | 0.07 |
| ASP178@CG-HIS177@ND1.HD1 | 13.85 | 23.91 | 0.40 | 15.38 | 14.53 | 12.28 | 29.15 | 20.82 | 21.22 |
| GLU211@CD-HIS177@ND1.HD1 | 8.29 | 4.67 | 88.56 | 23.20 | 24.23 | 7.75 | 24.53 | 8.77 | 7.85 |
| GLU196@CD-ARG156@CA.CZ | 5.51 | 10.04 | 16.45 | 65.75 | 0.86 | 42.63 | 0.05 | 28.88 | 36.37 |
| GLU186@CD-HIS187@ND1.HD1 | 5.13 | 0.81 | 0.29 | 7.95 | 26.29 | 1.13 | 80.43 | 5.88 | 9.35 |
| HIS187@CG-LYS185@CA.NZ | 2.03 | 0.13 | 0.62 | 0.16 | 1.18 | 0.32 | 0.57 | 1.20 | 3.63 |
| ASP202@CG-ARG156@CA.CZ | 1.69 | 2.19 | 21.40 | 5.68 | 3.33 | 1.07 | 0.50 | 0.67 | 2.10 |
| GLU207@CD-HIS177@ND1.HD1 | 0.77 | 1.70 | 6.87 | 6.35 | 5.36 | 1.39 | 6.30 | 1.32 | 1.58 |
| ASP144@CG-HIS140@ND1.HD1 | 0.22 | 1.53 | - | 0.64 | 0.83 | 10.14 | 17.32 | 10.10 | 0.32 |
| HIS155@NE2-ARG136@CA.CZ | 0.18 | 3.15 | 0.15 | 0.53 | 0.19 | 0.61 | 3.63 | 3.53 | 0.12 |
| HIS155@CG-ARG136@CA.CZ | - | 0.29 | - | 0.05 | 0.01 | 0.07 | 0.50 | 0.88 | |
| HIS140@NE2-ARG208@CA.CZ | 0.17 | - | - | - | - | 0.03 | - | 4.02 | 19.77 |
| HIS155@NE2-ARG151@CA.CZ | 0.14 | 0.83 | 0.97 | - | 0.10 | 0.01 | 38.38* | 20.33 | 32.00 |
| HIS155@CG-ARG151@CA.CZ | - | 0.04 | 0.01 | - | 0.01 | - | 49.22* | 18.75 | 41.98 |
| GLU196@CD-HIS155@ND1.HD1 | 0.06 | - | 0.51 | 11.34 | 0.17 | 6.62 | 0.05 | 2.35 | 4.05 |
| GLU196@CD-HIS187@ND1.HD1 | - | 0.03 | 0.75 | 0.08 | 0.01 | 0.15 | 0.28 | 1.07 | 1.40 |
| HIS187@NE2-HIS155@ND1.HD1 | 0.01 | - | 0.05 | 0.25 | 0.07 | 0.83 | 0.02 | 0.05 | - |
| GLU152@CD-HIS155@ND1.HD1 | 0.01 | 4.61 | - | 0.42 | 0.45 | 0.01 | 30.00* | 35.52 | 30.53 |
| GLU146@CD-LYS204@CA.NZ | 0.01 | - | 2.18 | 0.57 | 0.02 | 0.01 | - | - | - |
| GLU200@CD-LYS204@CA.NZ | - | 0.07 | - | - | 0.03 | 0.09 | 2.12 | 0.32 | 0.23 |
| HIS155@NE2-LYS194@CA.NZ | - | - | 0.88 | 3.34 | 2.05 | - | 0.03 | 1.63 | 0.23 |
| GLU211@CD-HIS140@ND1.HD1 | - | - | - | 0.25 | - | 0.01 | - | 1.93 | 0.48 |
| HIS140@NE2-ARG136@CA.CZ | - | - | - | 0.17 | - | - | 5.18 | 4.73 | 5.00 |
| HIS155@CG-LYS194@CA.NZ | - | - | - | 0.16 | 0.17 | - | - | - | 0.33 |
| HIS140@NE2-ARG151@CA.CZ | - | - | - | 0.15 | 0.01 | 0.03 | 5.60 | 1.52 | 8.05 |
| HIS187@NE2-LYS194@CA.NZ | - | - | - | 0.04 | - | - | 1.60 | 0.03 | 2.50 |
| GLU146@CD-HIS140@ND1.HD1 | 0.06 | - | - | - | - | - | 1.93 | 2.92 | 22.98 |
| ASP202@CG-LYS204@CA.NZ | - | 0.01 | - | - | - | - | 1.68 | 0.02 | - |
| Buffalo PrP / | - | 300 K | - | - | 350 K | - | - | 450 K | - |
|---|---|---|---|---|---|---|---|---|---|
| Salt Bridges (SBs) | Seed1 | Seed2 | Seed3 | Seed1 | Seed2 | Seed3 | Seed1 | Seed2 | Seed3 |
| GLU146@CD-ARG151@CA.CZ | - | - | - | - | - | - | 25.58 | 13.33 | 1.33 |
| HIS140@CG-ARG151@CA.CZ | - | - | - | - | - | - | 6.47 | 2.70 | 4.92 |
| HIS140@CG-ARG148@CA.CZ | - | - | - | - | - | - | 0.17 | 0.02 | 0.08 |
| HIS140@NE2-ARG148@CA.CZ | - | - | - | - | - | - | 1.18 | 0.03 | 0.22 |
| HIS187@CG-LYS194@CA.NZ | - | - | - | - | - | - | 1.52 | - | 0.32 |
| GLU146@CD-ARG148@CA.CZ | - | - | - | - | - | - | 16.88 | 3.52 | - |
| HIS140@CG-HIS155@ND1.HD1 | - | - | - | - | - | - | 8.57 | 0.02 | 1.85 |
| HIS140@NE2-HIS155@ND1.HD1 | - | - | - | - | - | - | 5.82 | 0.02 | 1.93 |
| HIS140@NE2-ARG156@CA.CZ | - | - | - | - | - | - | 2.08 | - | - |
| HIS140@CG-ARG156@CA.CZ | - | - | - | - | - | - | 1.25 | - | - |
| HIS155@CG-HIS140@ND1.HD1 | - | - | - | - | - | - | 8.03 | 0.02 | 1.93 |
| HIS155@NE2-HIS140@ND1.HD1 | - | - | - | - | - | - | 7.78 | 0.03 | 2.47 |
| ASP202@CG-LYS194@CA.NZ | - | - | - | - | - | - | 6.57 | - | - |
| GLU152@CD-ARG156@CA.CZ | - | - | - | - | - | - | 6.27 | 5.30 | 5.30 |
| ASP147@CG-LYS194@CA.NZ | - | - | - | - | - | - | 5.15 | - | - |
| ASP147@CG-LYS204@CA.NZ | - | - | - | - | - | - | 1.80 | - | - |
| ASP147@CG-ARG208@CA.CZ | - | - | - | - | - | - | - | 0.03 | - |
| GLU207@CD-LYS185@CA.NZ | - | - | - | - | - | - | 3.48 | - | 0.02 |
| GLU152@CD-HIS140@ND1.HD1 | - | - | - | - | - | - | 3.28 | - | - |
| GLU152@CD-ARG136@CA.CZ | - | - | - | - | - | - | - | 0.18 | - |
| GLU152@CD-LYS194@CA.NZ | - | - | - | - | - | - | 1.70 | - | 0.27 |
| GLU221@CD-ARG136@CA.NZ | - | - | - | - | - | - | 1.80 | 0.85 | - |
| GLU221@CD-HIS140@ND1.HD1 | - | - | - | - | - | - | - | 11.73 | - |
| GLU200@CD-LYS185@CA.NZ | - | - | - | - | - | - | 1.55 | - | - |
| GLU200@CD-HIS187@ND1.HD1 | - | - | - | - | - | - | 1.15 | - | - |
| HIS140@NE2-ARG220@CA.CZ | - | - | - | - | - | - | 0.93 | - | - |
| HIS140@CG-ARG220@CA.CZ | - | - | - | - | - | - | 0.63 | - | - |
| GLU186@CD-ARG156@CA.CZ | - | - | - | - | - | - | 0.60 | - | - |
| ASP202@CG-ARG148@CA.CZ | - | - | - | - | - | - | 0.42 | - | - |
| ASP144@CG-ARG208@CA.CZ | - | - | - | - | - | - | 0.27 | - | - |
| ASP144@CG-ARG151@CA.CZ | - | - | - | - | - | - | 0.23 | 1.83 | - |
| ASP144@CG-HIS155@ND1.HD1 | - | - | - | - | - | - | 0.18 | 0.25 | - |
| GLU200@CD-LYS194@CA.NZ | - | - | - | - | - | - | 0.17 | - | - |
| ASP167@CG-ARG164@CA.CZ | - | - | - | - | - | - | 0.15 | - | - |
| GLU196@CD-ARG148@CA.CZ | - | - | - | - | - | - | 0.13 | - | - |
| GLU196@CD-HIS140@ND1.HD1 | - | - | - | - | - | - | 0.03 | - | - |
| GLU146@CD-HIS155@ND1.HD1 | - | - | - | - | - | - | 0.10 | 0.08 | - |
| ASP202@CG-HIS140@ND1.HD1 | - | - | - | - | - | - | 0.07 | - | - |
| HIS177@NE2-ARG208@CA.CZ | - | - | - | - | - | - | 0.02 | - | - |
| HIS177@NE2-LYS185@CA.NZ | - | - | - | - | - | - | 0.02 | - | - |
| HIS140@CG-ARG208@CA.CZ | - | - | - | - | - | - | - | 2.85 | 6.77 |
| ASP202@CG-HIS187@ND1.HD1 | - | - | - | - | - | - | - | 1.07 | 3.02 |
| ASP147@CG-HIS155@ND1.HD1 | - | - | - | - | - | - | - | 0.02 | 0.02 |
| ASP147@CG-ARG136@CA.CZ | - | - | - | - | - | - | - | 0.22 | - |
| HIS187@NE2-LYS185@CA.NZ | - | - | - | - | - | - | 1.05 | - | - |
| ASP144@CG-ARG136@CA.CZ | - | - | - | - | - | - | - | 0.03 | - |
| HIS140@NE2-LYS204@CA.NZ | - | - | - | - | - | - | - | - | 0.03 |
| HIS155@NE2-ARG148@CA.CZ | - | - | - | - | - | - | - | 0.05 | - |
| GLU196@CD-LYS185@CA.NZ | - | - | - | - | - | - | 0.03 | - | - |
| GLU146@CD-ARG136@CA.CZ | - | - | - | - | - | - | 0.05 | 6.10 | - |
| GLU146@CD-ARG208@CA.CZ | - | - | - | - | - | - | - | 4.05 | 0.03 |
| HIS155@CG-HIS187@ND1.HD1 | - | - | - | - | - | - | - | - | 0.02 |
| HIS140@CG-ARG136@CA.CZ | - | - | - | - | - | - | 2.45 | 3.90 | 1.45 |
At 450 K, the SBs in H1 such as HIS155-ARG151, HIS155-GLU152 have been observed to have high occupied rates, and from Table 2, we may know that many low occupied rate SBs were not found at 300 K and 350 K. The removal of all these SBs under low environment will lead to the changes of H1 and H2 region of BufPrP from α-helices structures (BufPrPC) into β-sheet structures (BufPrPSc). Our further analyses of the B-factor and RMSF figures [27] can also provide confirmation of the findings from these SBs.
CONCLUSION
This brief article presents the MD results of BufPrP at different temperatures and provides a clue (from the salt bridge point of view) regarding the reasons for the conversion from normal cellular prion protein (PrPC) to diseased infectious prions (PrPSc). This should be very useful with respect to the goals of medicinal chemistry in prion diseases research fields.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
This research (with the project no. pb04 at Federation University Australia and under NCI) was undertaken with the assistance of resources and services provided by the National Computational Infrastructure (NCI), which is supported by the Australian Government.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The author thanks reviewers of this work for their valuable comments and suggestions that helped improve the presentation of this research work.


