Meta-Analysis of Expression of the Stress Tolerance Associated Genes and Uncover their Cis-Regulatory Elements in Rice (Oryza sativa L.)
Abstract
Background:
Rice contributes to the staple food of more than half of the world’s population. However, its productivity is influenced by various biotic and abiotic stresses. Genetic engineering and plant breeding tools help to overcome the adverse effects of environmental stresses. The advanced bioinformatics tools provide information for a better understanding of the mechanisms underlying stress tolerance, gene expression profiles and functions of the important genes and cis-regulatory elements involved in better performance under abiotic stresses.
Objective:
To identify the key genes involved in the tolerance mechanism for abiotic stresses and their regulatory networks in rice (Oryza sativa L.).
Methods:
A total of 152 various microarray datasets associated with nine rice trials were retrieved for expression meta-analysis through various bioinformatics tools.
Results:
The results indicated that 29593, 202798, 73224 and 25241 genes represented significant differential expression under cold, drought, salinity and heat stress conditions compared with the control condition, respectively. Twenty three highly overexpressed genes were identified under the evaluated abiotic stresses. The transcription regulatory activity of differentially expressed genes was mainly due to hormone, light and stress-responsive cis-acting regulatory elements among which ABRE, ARE, CGTCA-motif, GARE-motif, TGACG-motif, G-box, G-Box, GAG-motif, GA-motif, TCT-motif, Box 4, Sp1, HSE, MBS and TC-rich repeats were the most important in the promoter sites of the identified up-regulated genes. The results of cis-acting regulatory analysis suggest that 15 cis-acting regulatory elements were contributed to the tolerance mechanisms for abiotic stresses.
Conclusion:
The result of expression meta-analysis in this study provides an insight for plant breeders for better understanding the function of the genes and their regulatory mechanism in plants (especially cereals) exposed to different abiotic stresses. The outcome of this study suggests practical approaches for designing unified breeding programmes to breed multi-abiotic stress-tolerant species.
1. INTRODUCTION
Agriculture is affected by environmental abiotic stresses including drought, high salinity, low and high temperatures. Responses of plants to abiotic stresses are being increasingly addressed on a genome-wide scale in order to find novel gene targets involved in the tolerance mechanisms [1]. The share of rice to the diet of almost half of the worlds’ population and its popularity as a post-genomic model crop has made it an important crop for meta-analysis of stress tolerance associated genes [2, 3]. The response of gene families to abiotic stresses has been assessed by transcriptome-wide analyses suggesting their role in response to multiple environmental stresses [4-7]. Genetic analysis of the tolerance mechanisms against environmental stresses in plants reveals that specific genes respond to such stresses at the transcriptional level [8-15]. At the molecular level, environmental stresses affect the expression of stress-responsive genes [16-21]. The products of the stress-inducible genes have been classified into two classes: one that directly protects against environmental stresses and the other that is associated with gene expression and signal transduction pathways. Detection of stress-inducible genes helps to improve the stress tolerance in plants through cross-breeding and genetic engineering tools [8-10, 14, 15, 22-24]. Identification of stress-inducible genes and expression changes in response to environmental stresses and a better understanding of the regulatory mechanism behind differentially expressed genes are important for the improvement of crop plants under abiotic stress conditions.
Regulation of gene expression in various tissues during physiological processes is controlled at the transcriptional, post-transcriptional and post-translational levels. Regulation at the transcriptional level that plays an important role in response to abiotic stresses is mainly associated with promoters and their contributing cis-acting regulatory elements (CARE) [25]. Promoters are DNA sequences located at the upstream of the gene coding region and included the CAREs as the binding sites for proteins in transcription events. In higher plants, CAREs act as enhancers, silencers or insulators [26]. Promoters have a core segment of 40 bp at the upstream of the transcription initiation site that compromises the TATA-box [27, 28]. Proximal and distal regions located at the upstream of the core promoters (regulatory sequence and cis-elements) play a significant role in the regulation of gene expression [28, 29]. Promoter analysis provides valuable information to identify the function and signalling of genes. Furthermore, CARE is the appropriate goal to dissect the molecular mechanisms of responses to abiotic stresses [30]. As a result, access to gene expression data helps to better understand the expression pattern of gene families or single genes at whole genome level and accommodates to identify the gene(s) contributed to biological processes [31]. DNA microarray representing high-throughput gene expression profiling has been used to discern the tolerance mechanisms in plants [32-41]. Besides, the gene expression databases and various bioinformatics tools help to determine cis-regulatory sites in coding and non-coding DNA sequences [42].
Analysis of expression patterns of stress responsive genes in rice associated trials resulted in a large volumes of expression data that are available as online databases. However, the exact molecular mechanism underlying stress responses is still poorly understood. Hence, there is a great demand to assess expression data through meta-analysis and identify commonly expressed genes under various abiotic stresses conditions that can be used in engineering stresses tolerance in rice. The aims of the present study were to (1) assess subsets of expression data associated with stress tolerance retrieved from online databases using bioinformatics tools; (2) meta-analysis of common expressed genes contributing to cold, heat, drought and salinity tolerance in rice; and to (3) uncover and characterize the regulatory mechanisms of abiotic tolerance responsive genes (Table 1).
2. METHODS
2.1. Database Development and Expression Meta-Analysis
A database containing 152 microarray expression data sets corresponding to nine rice trials associated with cold, salt, drought and heat-stressed conditions was developed (Supplementary Table 1). The rice microarray data sets were retrieved from the ArrayExpress and NCBI GEO DataSets [43, 44]. The expression data have been annotated to the ArrayExpress and NCBI GEO DataSets. All the data sets, title, experiment type and the overall design for downloading the data can be found on the ArrayExpress (https://www.ebi.ac.uk/ arrayexpress/) and NCBI (http://www.ncbi.nlm.nih.gov/) documentation pages [45-53]. Furthermore, the library (.CDF: rice_libraryfile) and annotation (.CSV: Rice.na35.annot.csv) files were retrieved from the Affymetrix database (http://www.affymetrix.com/technology/mip_technology.affx). Selection of these data files was based on the type of Chips that have been used in microarray experiments. The Affymetrix raw data (.CEL) files were analyzed by FlexArray software version 1.6.3. Firstly, the CEL data and the library file were imported to the FlexArray software. The CEL data were normalized based on the Robust Multiarray Average (RMA) algorithm [54] and the RMA signal values were transformed into Log2. The RMA is a preprocessing algorithm used for background correction and data normalization in Affymetrix and Nimblegen gene expression microarray trials. In order to identify Differentially Expressed Genes (DEGs), the RMA expression values were analyzed based on a two-sample student’s t-test. Finally, the gene list of the expression data identified using t-test and the annotation for each gene were added based on a new rice annotation file. The up-regulated genes with above 1- symmetrical raw fold change (FC), the down-regulated genes of less than 0-symmetrical raw FC and the over-expressed genes with above 30- symmetrical raw FC were selected for further data analysis (Table 2). The P-values were adjusted in the false discovery rate (FDR) of less than 0.05.
2.2. Differentially Expressed Genes (DEGs)
To compare the results of expression data analyses, the UpSetR R package was used to identify the interrelationship between the DEGs and the representing interactions among the gene sets identified under cold, heat, salinity and drought stress conditions.
2.3. Gene Ontology (GO) and Functional Categorization
Annotations of gene for overexpressed genes and clus-tering functions, location and biological roles were accomp-lished by the GO Tutorial-TAIR at https://www.arabidopsis. org/help/tutor-ials/go6.jsp. Furthermore, the GO annotation and the functional categorizations derived from the TAIR (https://www.arabidopsis.org/tools/bulk/go/index.jsp) database. Functional categorization was performed followed by the below equation:
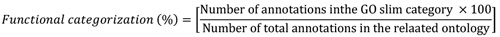 |
| Condition | Probeset | Gen Bank Acc. | Fold Change | T Statistic | P-value |
|---|---|---|---|---|---|
| Cold Stress | Os.52451.1.A1_at | AK067195.1 | 83.19 | 11.17 | 3.60E-04 |
| Os.32366.1.S1_at | AK105196.1 | 78.70 | 15.90 | 1.99E-08 | |
| Os.52280.1.S1_at | AK066054.1 | 33.45 | 33.07 | 4.98E-06 | |
| Drought Stress | Os.25497.1.S1_at | CA765994 | 955.99 | 306.68 | 1.06E-05 |
| Os.51718.1.S1_at | AK063517.1 | 926.56 | 36.38 | 3.41E-06 | |
| Os.47732.1.S1_at | BI809490 | 797.08 | 61.51 | 2.60E-04 | |
| Os.49245.1.S1_at | AK063685.1 | 746.08 | 74.90 | 1.90E-07 | |
| Os.5325.1.S1_at | AK107930.1 | 738.38 | 20.21 | 3.54E-05 | |
| Os.8668.1.S1_x_at | AK066459.1 | 637.73 | 163.33 | 3.75E-05 | |
| Os.12551.1.S1_s_at | U57641.1 | 585.38 | 142.02 | 1.47E-08 | |
| Os.12633.1.S1_s_at | U60097.2 | 553.39 | 41.28 | 1.35E-08 | |
| Os.42784.1.S1_at | NM_189885.1 | 521.36 | 22.12 | 5.58E-07 | |
| Os.28200.1.S1_x_at | AK099709.1 | 511.55 | 39.61 | 1.73E-08 | |
| Os.37717.1.A1_s_at | BU673746 | 473.89 | 6.87 | 2.30E-03 | |
| Os.11271.2.S1_at | CB666821 | 374.63 | 138.28 | 5.23E-05 | |
| Os.12415.1.S1_at | AK063582.1 | 335.29 | 44.93 | 4.90E-04 | |
| Os.11260.1.S1_at | AK102039.1 | 156.35 | 237.82 | 1.77E-05 | |
| Os.47625.1.A1_s_at | BX901098 | 117.62 | 8.18 | 1.40E-02 | |
| Salt Stress | Os.49245.1.S1_at | AK063685.1 | 413.38 | 62.04 | 4.04E-07 |
| Os.23092.1.S1_at | CA755805 | 319.17 | 126.75 | 6.22E-05 | |
| Os.5325.1.S1_at | AK107930.1 | 154.66 | 32.22 | 5.53E-06 | |
| Os.12633.1.S1_s_at | U60097.2 | 100.73 | 26.57 | 1.19E-05 | |
| Os.56004.1.S1_at | AK109114.1 | 99.61 | 29.87 | 1.22E-08 | |
| Os.12415.1.S1_at | AK063582.1 | 81.91 | 26.16 | 1.27E-05 | |
| Os.12703.1.S1_at | AK070417.1 | 63.08 | 15.75 | 1.01E-06 | |
| Os.51718.1.S1_at | AK063517.1 | 62.40 | 24.61 | 1.62E-05 | |
| Heat Stress | Os.11039.3.S1_at | AK105370.1 | 661.77 | 193.18 | 4.31E-09 |
| Os.11039.1.S1_s_at | AK063751.1 | 619.97 | 75.44 | 1.85E-07 | |
| Os.47625.1.A1_s_at | BX901098 | 112.82 | 7.19 | 1.18E-02 | |
| Os.10038.1.S1_s_at | AU082861 | 100.98 | 10.83 | 8.40E-03 |
| Cis-Regulatory Element | Sequence | Function |
|---|---|---|
| AAGAA-motif | GAAAGAA | Unknown |
| A-box | CCGTCC | Cis-acting regulatory element |
| ABRE | TACGTG | Cis-acting element involved in the abscisic acid responsiveness |
| ACE | AAAACGTTTA | Cis-acting element involved in light responsiveness |
| ARE | TGGTTT | Cis-acting regulatory element essential for the anaerobic induction |
| Box 4 | ATTAAT | Part of a conserved DNA module involved in light responsiveness |
| CAAT-box | CAAT | Common cis-acting element in promoter and enhancer regions |
| CAT-box | GCCACT | Cis-acting regulatory element related to meristem expression |
| CCGTCC-box | CCGTCC | Cis-acting regulatory element related to meristem specific activation |
| CGTCA-motif | CGTCA | Cis-acting regulatory element involved in the MeJA-responsiveness |
| Circadian | CAANNNNATC | Cis-acting regulatory element involved in circadian control |
| GAG-motif | GGAGATG | Part of a light responsive element |
| GA-motif | AAAGATGA | Part of a light responsive element |
| G-box | TACGTG | Cis-acting regulatory element involved in light responsiveness |
| G-Box | CACGTA | Cis-acting regulatory element involved in light responsiveness |
| HSE | AAAAAATTTC | Cis-acting element involved in heat stress responsiveness |
| MBS | TAACTG | MYB binding site involved in drought-inducibility |
| O2-site | GATGATATGG | Cis-acting regulatory element involved in zein metabolism regulation |
| SKn-1-motif | GTCAT | Cis-acting regulatory element required for endosperm expression |
| Sp1 | CC(G/A)CCC | Light responsive element |
| TATA-box | TATAAA | Core promoter element around -30 of transcription start |
| TC-rich repeats | ATTTTCTTCA | Cis-acting element involved in defense and stress responsiveness |
| TCT-motif | TCTTAC | Part of a light responsive element |
| TGACG-motif | TGACG | Cis-acting regulatory element involved in the MeJA-responsiveness |
Where, GO slims (subsets) are cut-down versions of the gene ontology containing a subset of the terms. This structure represents a subset of the ontology that was designed specifically for plants and can be used for organizing sets of genes according to broad GO ontology categories. The nominator of the equation represents annotation count of a specific functional category (nucleus, cell, nuclease activity) in each GO category (cellular component, molecular function or biological process) and the denominator stands for the total annotation count of the functional category in each GO category.
2.4. Analysis of Cis-Acting Regulatory Element (CAREs)
The accession numbers of the complete nucleotide sequence (FASTA format) of the over-expressed genes contributing to transcription regulatory activities were obtained from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) database. For CAREs analysis, the 1.5 kbp sequences of 5´ upstream of each gene were retrieved from the Phytozome (http://www.phytozome. net/) database and subsequently were subjected to the PlantCare (http://bioinformatics.psb.ugent.be/webtools/plant care/html/) database to identify common CAREs in the responsive genes against the abiotic stresses [55].
3. RESULTS AND DISCUSSION
3.1. Identification of DEGs
The results of the t-test in meta-analysis suggested 29593, 202798, 73224 and 25241 gene accessions with significant differential expression under cold, drought, salinity and heat stresses, respectively (Supplementary Table 2). These data suggested that drought and salinity stresses were more restrictive than cold and heat stresses for rice growth.
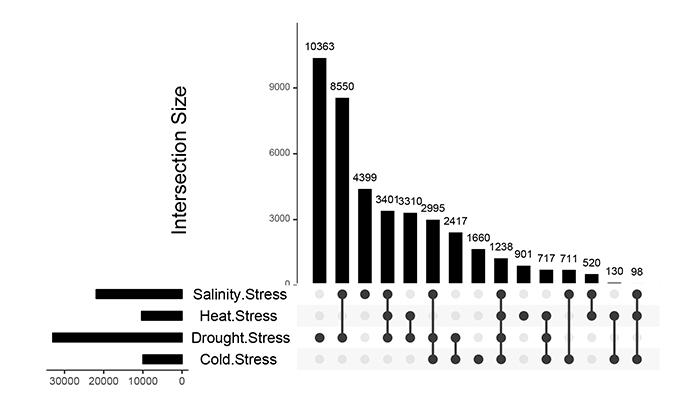
Differences and cross-talk of gene expression among drought, salinity, cold and heat stress responses were analyzed using Upset plots. As shown in Fig. (1), 10363, 4399, 1660 and 901 gene accessions represented up-regulation under drought, salt, cold and heat stresses, respectively. Furthermore, the expression of 1238 genes in the four stresses was increased. A number of 8550, 2417, 520 and 130 genes showed an increase in expression level under drought-salinity, drought-cold, salinity-heat and cold-heat stress conditions, respectively (Fig. 1) suggesting the existence of greater crosstalk between drought and salinity stress signaling processes in rice.
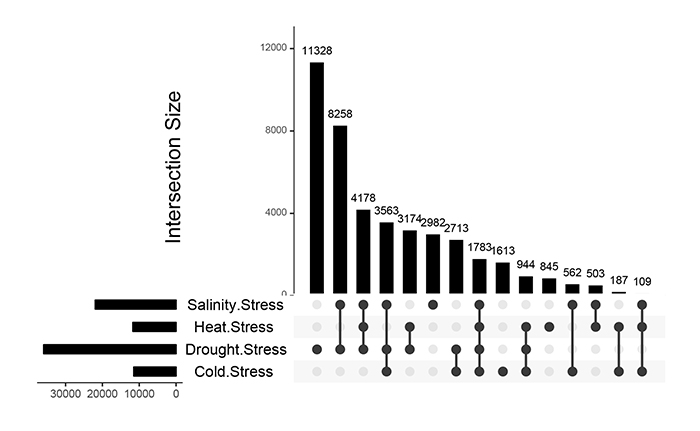
Analysis of down/up-regulated stress related genes helps a better understanding of the basis of molecular responses to abiotic stresses [38]. In the present study, the results of the meta-analysis revealed that a total of 11328, 2982, 1613 and 845 gene accessions were down-regulated under drought, salinity, cold and heat stresses (Fig. 2). The results indicated that 8258, 2713, 503 and 187 DEGs showed crosstalk in response to drought-salinity, drought-cold, salinity-heat and cold-heat stress combinations, respectively. The number of down-regulated genes was higher than that of up-regulated showing the majority of genes have been switched off under drought and salinity stresses in rice (Figs. 1 and 2). Similar results were obtained in arabidopsis tested under stressed conditions [38].
Of the up-regulated genes, 47% were associated with potential candidate genes contributed to abiotic stress tolerance. Some of the key genes involved in stress-induced proteins, brassinosteroid-regulated proteins, binding proteins, pathogenesis-related proteins, transcription factors, photo-synthetic proteins and transporter proteins. The up-regulation of these genes under drought, cold and salinity stresses has been reported previously [38]. A number of 24 genes over-expressed in various environmental conditions were selected for further analysis to provide a valuable resource of information for use in breeding programs under abiotic stresses (Table 1).
3.2. GO and Functional Annotation of DEGs
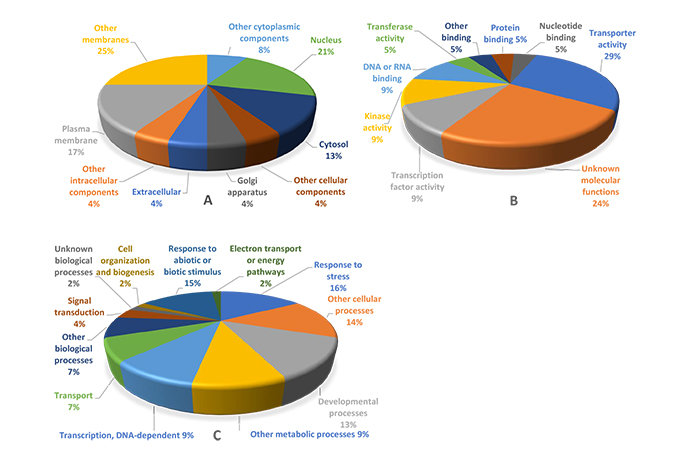
Functional groups of overexpressed genes in the four stresses tested are presented in Fig. (3). The results of the functional annotation of DEGs showed that the nucleus, other membranes and plasma membrane gene groups had the highest percentages in cellular component category. Several genes with specific products in certain places of cell (cell membranes) showed up-regulation in response to abiotic stresses. The results of gene expression analysis revealed that trehalose that protects membranes and proteins in cells exposed to drought stress conditions was accumulated under various abiotic stresses [56, 57].
Transporter activity, DNA and RNA-binding, transcription factor activity and kinase activity were the most prevalent gene groups among the identified over-expressed genes (Fig. 3) demonstrating their roles in alleviating the adverse effects of abiotic stress conditions in rice. RNA-binding proteins have an important role in post-transcriptional gene regulation. Most of the RNA-binding proteins are plant-specific with known functions. RNA-binding proteins that regulate pre-mRNA splicing, polyadenylation, RNA stability and RNA export are important for the adaptation of plants to various environments [58, 59]. The functional gene group for DNA binding activity contributes to tolerance to multiple stresses, generally in an ABA-independent manner through DRE/CRT cis-elements and the AP2/ERF DNA binding domain [60].
Various products of genes with kinase activities (Fig. 3) contribute to abiotic stress tolerance. Among the protein kin-ases involved in stress signal transduction, mitogen-activated protein kinases (MAPKs) [61-63], glycogen synthase kinase 3 (GSK3) [64, 65], S6 kinase (S6K) [55], calcium-dependent protein kinases (CDPKs) [66-68] and most of the SNF1-related kinases (SnRKs) are common among all eukaryotic organisms [69]. Furthermore, SnRK2 family members are plant-specific serine/threonine kinases contributed to plant response to abiotic stresses and abscisic acid (ABA)-dependent plant development [69].
Some of the identified overexpressed genes belonged to the Transcription Factors (TF) category (Fig. 3). It has been shown that TFs belonging to the dehydration-responsive element-binding proteins (DREB), C-repeat-binding factors (CBF), ABA-Binding Factors (ABF), myelocytomatosis oncogenes (MYC) and myeloblastosis oncogenes (MYB) respond to drought stress conditions [70-72]. Genes involved in kinase activity help plants deal with abiotic stresses. One of the most important plant TFs is DREB that regulates the expression of many stress-inducible genes mostly in an ABA-independent manner. The DREBs play a critical role in improving the abiotic stress tolerance of plants by interacting with a DRE/CRT cis-element present in the promoter regions of various abiotic stress-responsive genes [60]. The DREB TFs contain a highly conserved AP2/ERF DNA-binding domain across the plant kingdom including arabidopsis, rice, soybean, chickpea, tomato, tobacco, and millets [73]. Busk and Pages [74] also reported that phosphorylation is necessary for the activation of proteins under drought-stress conditions, thus enhancing the DNA-binding activity of several transcription regulators. Furthermore, a large portion of biological processes of the overexpressed genes under abiotic stresses was related to response to stress, response to abiotic and biotic stimuli, developmental processes and other cellular processes (Fig. 3). These results suggest the role of key genes responsible for the regulation of the most important biological processes under various abiotic stress conditions.
3.3. Identification of CAREs in the Promoter Region of Transcription Factors
The detected CAREs at the upstream of the DEGs under drought, heat, cold and salinity stress conditions were associated with a light response, hormonal regulation and stress responses in rice (Table 2 and Fig. 4). The hormonal regulatory elements included methyl jasmonate (MeJA) and abscisic acid (ABA) responsive motifs such as ABRE, CGTCA-motif and TGACG- elements were presented in the majority of the DEGs identified under salinity, cold, heat and drought stress conditions (Table 2).
Phytohormones play a key role in response to prioritization stresses [75]. Phytohormone signaling mediated by ABA is an evolutionarily conserved mechanism that promotes abiotic stress tolerance in plants [76]. The process of plants perceives, response and adaptation to abiotic stresses are controlled mainly by ABA that regulates plant water situation as an endogenous messenger [77]. Abscisic acid is a plant stress hormone because it induces under various stresses [78, 79]. Induction of the ABA hormone often relies on the presence of a cis-acting element called ABRE (ABA-responsive) element [8, 10, 80]. The ABRE elements play a key role in abscisic acid response that induced abiotic stresses, seed dormancy and maturation processes [1]. The ABRE elements are one of the most important CRE in rice and located near the transcription start site (TSS) [81, 82].
Both MeJA and Jasmonic acid (JA) contribute to a wide range of environmental conditions and physiological events comprising of seed germination and leaf senescence [83]. Jasmonic acid participates in plant growth and plays critical roles in both biotic and abiotic stress responses [84-86]. The application of MeJA alleviates the adverse effects of environmental stresses [87]. The MeJA suppresses the absorption of toxic ions, and reduces the adverse effects of osmotic stress through regulating inorganic penetrating ions or organic [87]. The results of the current study suggest that TGACG and CGTCA motifs are involved in MeJA response and regulation of plant defense against abiotic stresses. The MeJA activates antioxidant systems to detoxify Reactive Oxygen Species (ROS) in stressed plants [87, 88]. It has been shown that the signaling networks related to ABA and JA hormones are correlated [89].
The light-responsive elements (LREs) including ACE, BOX4, GAG-motif, G-box, G-Box, GA-motif, TCT-motif and Sp1 were identified in the promoter regions of the expressed salinity, cold, heat and drought-responsive genes (Table 2). These regulatory elements play a critical role in the regulation of transcriptional activity [90]. The LREs such as G-box, Box-4, GAG, GAP, GA motifs have been identified in the regulatory regions of the light-regulated genes that are needed for light-controlled transcriptional activities [91, 92]. The role of the G-Box element in arabidopsis; as part of the response mechanism against abiotic stresses has been uncovered. The role of GAG motif and G-Box element in response to abiotic stresses has been reported in studies with tobacco and wheat [93, 94].
A circadian element was detected in the drought, cold, heat and salinity responsive genes (Table 2). The circadian clock coordinates the responses of plants to multiple environmental challenges. The results of the present study indicated that the circadian clock may reinforce the plant’s ability to reduce the adverse effects of abiotic stresses. Results of transcriptomic analyses have shown that circadian clock controls several genes associated with response to salinity, drought and cold stresses [95, 96].
The stress-responsive elements such as TC-rich repeats, heat shock elements (HSE) and the myeloblastosis binding sequence (MBS) were presented in the majority of responsive genes (Table 2). The role of TC-rich repeats in responses to environmental stresses has been documented in previous reports [97-100]. Heat shock element is a transcription factor that binds to HSE cis-acting elements in the promoter of stress-inducible genes and plays central roles in the acquisition of plant tolerance against abiotic stresses [101]. MBS, which is a binding site for MYB transcription factors, controls many abiotic stress responses [102, 103]. Molecular approaches discerned the functional characterization of MYB domain proteins, particularly the R2R3-type members in various plant species, including rice, maize and soybean [103, 104]. A genome-wide comparative analysis of MYB genes and their expression in arabidopsis and rice suggested the potential role of MYB domain proteins in plant stress responses [105]. Several members of R2R3-type MYB transcription factors are involved in the regulation of the phenylpropanoid pathway and the production of various secondary metabolic compounds under abiotic stress conditions. The role of MBS elements against drought stress in common bean [106] and maize [107] has been previously reported. Furthermore, the effects of MYB TFs in low-temperature, light and osmotic stress induction responses have been uncovered [108-111].
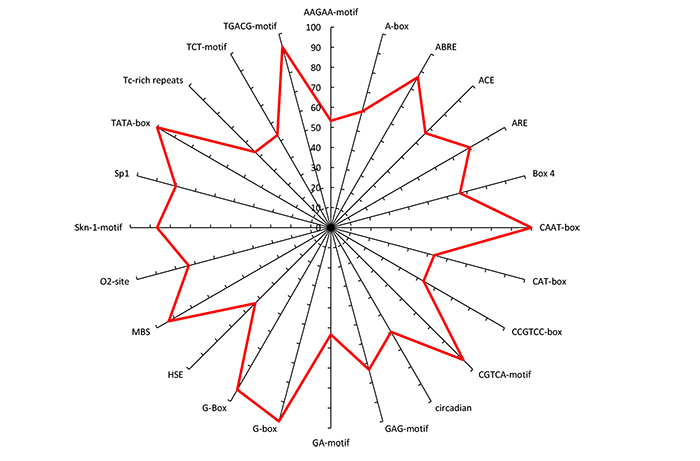
The results of the present study showed that ARE regulatory elements are essential for the induction of anaerobic respiration (Table 2). Moreover, the Skn-1 motif and the O2-site were present in the majority of responsive genes specifically those that act in the endosperm or involved in the zein metabolism regulation events. The role of these elements against abiotic stresses is restricted. However, the results of our study indicated the possibility of the role of these elements in tolerance against abiotic stresses in rice.
CONCLUSION
The expression pattern and regulation network of rice as a post-genomic crop under different abiotic stresses (heat, salt, cold and drought) were assessed on the basis of meta-analysis methods. The higher number of differentially expressed genes (DEGs) in drought and salinity environments compared to heat and cold stresses suggested greater crosstalk between drought and salinity stress signalling processes in rice. The higher number of co-expressed genes under drought and salinity stresses demonstrated strong correlations between the responses of rice to these stresses compared to the other cross-talks tested. Furthermore, 24 common over-expressed genes were identified in response to cold, drought, heat and salt stresses. These uncovered genes had DNA and RNA-binding role, transcription factor activity and kinase activity that were associated with both abiotic and biotic stimuli. Moreover, the identified CAREs at the upstream of common DEGs under drought, heat, cold and salinity stress conditions were related to light response, hormonal regulation and stress-related responses in rice. Overall, the outcome of this study helps to better understand the regulation mechanisms of stress responses at the transcriptional level.
LIST OF ABBREVIATIONS
| RMA algorithm | = Robust Multiarray Average Algorithm |
| DEGs | = Differentially Expressed Genes |
| CBF | = C-repeat-Binding Factor |
| GO | = Gene Ontology |
| FDR | = False Discovery Rate |
| MAPKs | = Mitogen-Activated Protein Kinases |
| NCBI | = National Center for Biotechnology Information |
| DREB | = Dehydration Responsive Element Binding |
| CAREs | = Cis-Acting Regulatory Element |
| MeJA | = Methyl Jasmonate |
| GA | = Gibberellin |
| GSK3 | = Glycogen Synthase Kinase3 |
| CDPKs | = Calcium-Dependent Protein Kinases |
| S6K | = S6 Kinase |
| SnRKs | = SNF1-Related Kinases |
| ABA | = Abscisic Acid |
| ABF | = Abscisic Acid-Binding Factor |
| MYC | = Myelocytomatosis oncogene |
| MYB | = Myeloblastosis Oncogene |
| TFs | = Transcription Factors |
ETHICS APPROVAL AND CONSENT TO PARTI-CIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
STANDARD OF REPORTING
PRISMA Guideline and methodology were followed.
AVAILABILITY OF DATA AND MATERIALS
The data supporting the findings of the article is available in the Dropbox cloud storage at: https://www.dropbox.com/s/ qlz9ybgoka2dyt2/Rice%20Raw%20Data_Library%20and%20Annotation%20File.rar?dl=0.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the support from Shiraz University.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers web site along with the published article.


