All published articles of this journal are available on ScienceDirect.
ACE2 Shedding and Furin Abundance in Target Organs may Influence the Efficiency of SARS-CoV-2 Entry
Abstract
Background:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a lineage B coronavirus, causing the worldwide outbreak of Corona Virus Disease 2019 (COVID-19). Despite genetically closed to SARS-CoV, SARS-CoV-2 seems to possess enhanced infectivity and subtle different clinical features, which may hamper the early screening of suspected patients as well as the control of virus transmission. Unfortunately, there are few tools to predict the potential target organ damage and possible clinical manifestations caused by such novel coronavirus.
Methods:
To solve this problem, we use the online single-cell sequence datasets to analyze the expression of the major receptor in host cells that mediates the virus entry, including angiotensin converting enzyme 2 (ACE2), and its co-expressed membrane endopeptidases.
Results:
The results indicated the differential expression of ADAM10 and ADAM17 might contribute to the ACE2 shedding and affect the membrane ACE2 abundance. We further confirm a putative furin-cleavage site reported recently in the spike protein of SARS-CoV-2, which may facilitate the virus-cell fusion. Based on these findings, we develop an approach that comprehensively analyzed the virus receptor expression, ACE2 shedding, membrane fusion activity, virus uptake and virus replication to evaluate the infectivity of SARS-CoV-2 to different human organs.
Conclusion:
Our results indicate that, in addition to airway epithelia, cardiac tissue and enteric canals are susceptible to SARS-CoV-2 as well.
1. INTRODUCITON
SARS-CoV-2 was distinguished as group 2B CoV and genetically related to SARS-CoV with ~80% nucleotide identity [1, 2]. Although patients with SARS-CoV-2-induced pneumonia exhibited typical symptoms of viral pneumonia, including cough, fever, myalgia, headache and even Acute Respiratory Distress Syndrome (ARDS), a substantial portion of patients could experience atypical symptoms like diarrhea (gastrointestinal discomforts), acute kidney injury, liver damage and acute cardiac injury [3], challenging clinical inter- ventions and long-term rehabilitations. Here, we speculated that in-depth analysis of virus entry into host cells would be crucial to explain the different sensitivities of various cell types to SARS-CoV-2 and the above-mentioned diversity of clinical manifestations.
Coronaviruses have been reported to enter host cells in two distinct routes [4]. The first route is the well-known receptor-mediated endocytosis. The surface spike (S) glycoprotein is widely accepted as the major determinant of coronaviruses entry. The S1 domain mediates the receptor binding and the S2 domain responsible for virus-cell membrane fusion [5]. Upon binding to the corresponding receptor on host cells surface, the S protein undergoes subtle conformational changes result in the uptake by endosomes for further proteolytic digest [6, 7]. In the case of SARS-CoV, numerous studies have demonstrated Angiotensin Converting Enzyme 2 (ACE2) receptor on the surface of human cells could be utilized to facilitate virus entry [8, 9]. Given close relation to SARS-CoV, emerging evidence indicates the S1 subunit of SARS-CoV-2 may also bind ACE2 receptors in lung, renal and cardiac tissue, liver, testis and intestinal epithelia to mediate the virus fusion and lead to the related symptoms [10-13]. Furthermore, ACE2 could be shed from the surface of human airway epithelia, the site of SARS-CoV infection, by a disintegrin and metalloproteinase (ADAM) family of proteases, which contributes to the ACE2 downre- gulation [14, 15]. This means the expression pattern of ACE2 in normal tissues may not directly correlate with their suscep- tibility to the SARS-CoV-2.
Alternatively, the S protein could be cleaved at the interface of the receptor binding (S1) and fusion (S2) domains (S1/S2), as well as in a novel position adjacent to a fusion peptide within S2 (S2′) by membrane endopeptidases co-exp- ressed with the host cell receptor, thereby inducing the direct fusion of viral and cellular membrane [16, 17]. Millet et al. identified the S protein of Middle East respiratory syndrome coronavirus (MERS-CoV) could be cleaved and activated by furin, a ubiquitously expressed endopeptidase, which is essen- tial for the MERS-CoV infection [17]. Additionally, transmem- brane protease, serine 2 (TMPRSS2) was reported to directly cleave and activate SARA-S, rendering SARS-S-driven virus-cell fusion [18, 19]. However, few researchers investigate the potential endopeptidases involved in the cleavage at the S protein of SARS-CoV-2, leading to the infections of multiple organs.
Based on the above, the distribution of ACE2 may not strictly be correlated with the cell tropism of SARS-CoV-2. Other influencing factors should be taken into consideration to predict the potential tissue damage besides a pulmonary injury. In the current study, we analyzed the online single-cell seque- nce datasets to unveil the expression pattern of ACE2 as well as ACE2 co-expressed membrane endopeptidases to comp- rehensively assess the infective efficiency of SARS-CoV-2 in different organs. Furthermore, we emphasized the overall analysis of ACE2 expression, FURIN expression and ACE2 shedding to explain the various clinical manifestations in different targeted organs after SARS-CoV-2 infection.
2. METHODS
2.1. Public scRNA-seq Dataset Acquisition
The gene expression and cell type annotation of single cell RNA-sequencing (scRNA-seq) data from normal human lung (GSE122960), nose (GSE121600), heart (GSE109816, GSE1- 21893) and enteric canal (GSE125970, GSE119969) were downloaded from the Gene Expression Omnibus.
2.2. scRNA-seq Data Processing
Seurat (version 2.3.2) and Scanpy (1.4.4.post1) [20, 21] were used to perform the scRNA-seq data processing following the standard procedure (mentioned athttps://scanpy.tutorials. readthedocs.io/en/latest/pbmc3k.html and https://satijalab.org/ seurat/v3.1/pbmc3k_tutorial.html)
2.3. Filtering of ACE2 Co-expressing Genes
All original data were treated through a normalization procedure by Scanpy (https://scanpy-tutorials.readthedocs.io/ en/latest/pbmc3k.html). Cell count matrixs were normalized in order to make them comparable through all qualified cells. The newly formed normalization matrixes were used to find genes co-expressing with ACE2.
Due to the limitation and noise of current scRNA-seq methods, the co-expressed gene-network is usually constructed using scRNA-seq data for grouped cells instead of each single cell [22, 23]. According to this strategy, cells with ACE2 expression >0 were selected to group as the subject of our study. From these filtered ACE2 positive cells, if the number of expressing cells about one gene did not exceed the 10 percent of the total ACE2 positive cells, this gene would be excluded. The rest would be regarded as ACE2 co-expressing genes.
2.4. Signature Scores
The signature scores were calculated basing on the method mentioned by A Wallrapp et al. [24], as follows.
Let S be a set of genes, we got
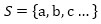 |
For a given cell, signature score can be calculated as:
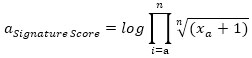 |
Where xi is the normalized gene expression, n is the total number of elements in the gene set.
The Violin plots representing the gene expressions and score values were drawn by seaborn (https://seaborn.pydata. org/) and matplotlib (https://matplotlib.org/).
2.5. Sequencing Analysis of Virus Spike Protein
The complete genome and protein sequences of SARS-CoV-2 (NC_045512.2) and SARS-Cov (NC_004718.3) were acquired from GenBank. The protein sequences of their spike protein were picked up for complete alignment with software Clustal X2 [25]. The secondary structure prediction of the spike protein of SARS-CoV-2 was performed by the online tool SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat. pl?page=npsa_sopma.html).
3. RESULTS
3.1. Candidate Endopeptidases Related to Virus Entry
The alveolar type II cells (SFTPC positive) of the lung were enrolled in our study. After filtering the ACE2 co-expressed genes, eight candidate endopeptidases related to virus entry were selected according to the selection flow chart (Fig. 1A), including ADAM10, ADAM17, FURIN, SPPL2B, TMPRSS2, HPN, CAPN1 and CAPNS1. They are all located on cell membrane and related to the virus entry of SARS-CoV-2 (Fig. 1B).
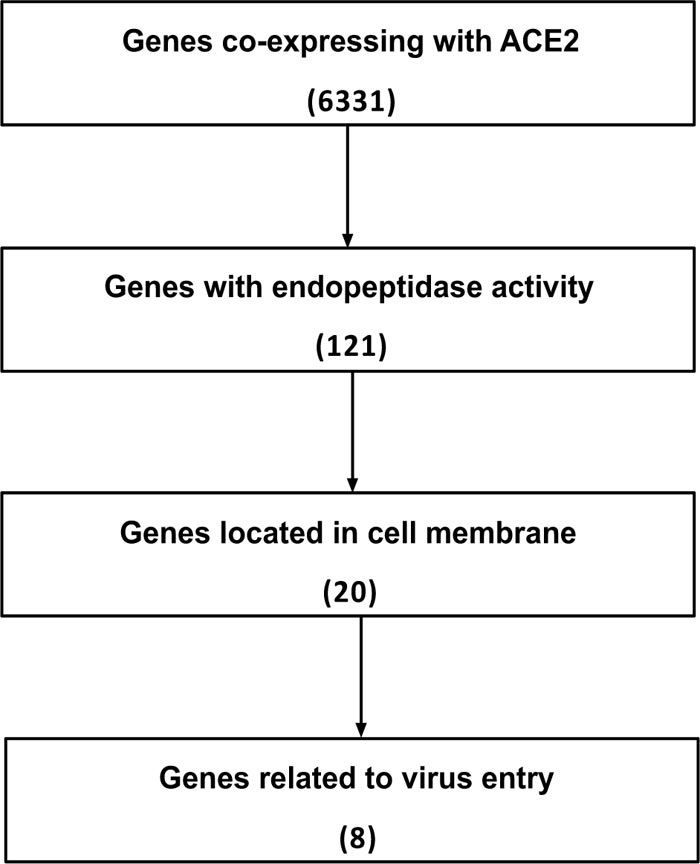
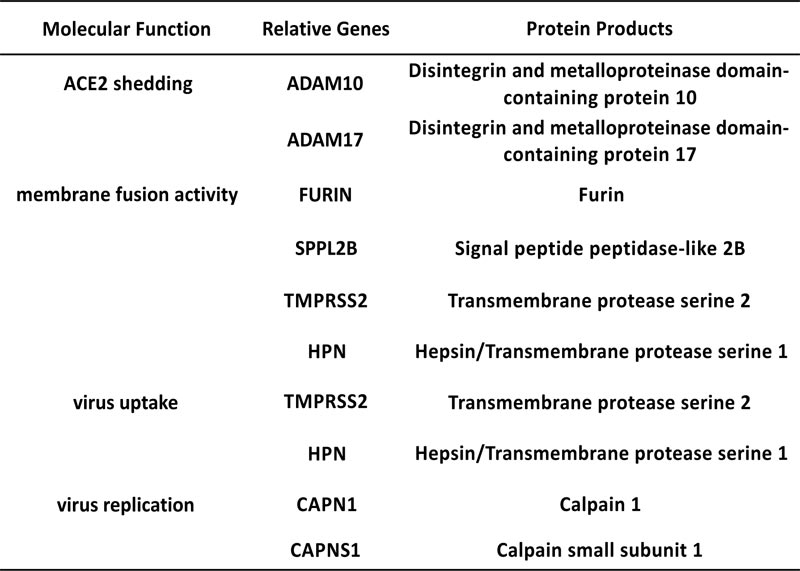
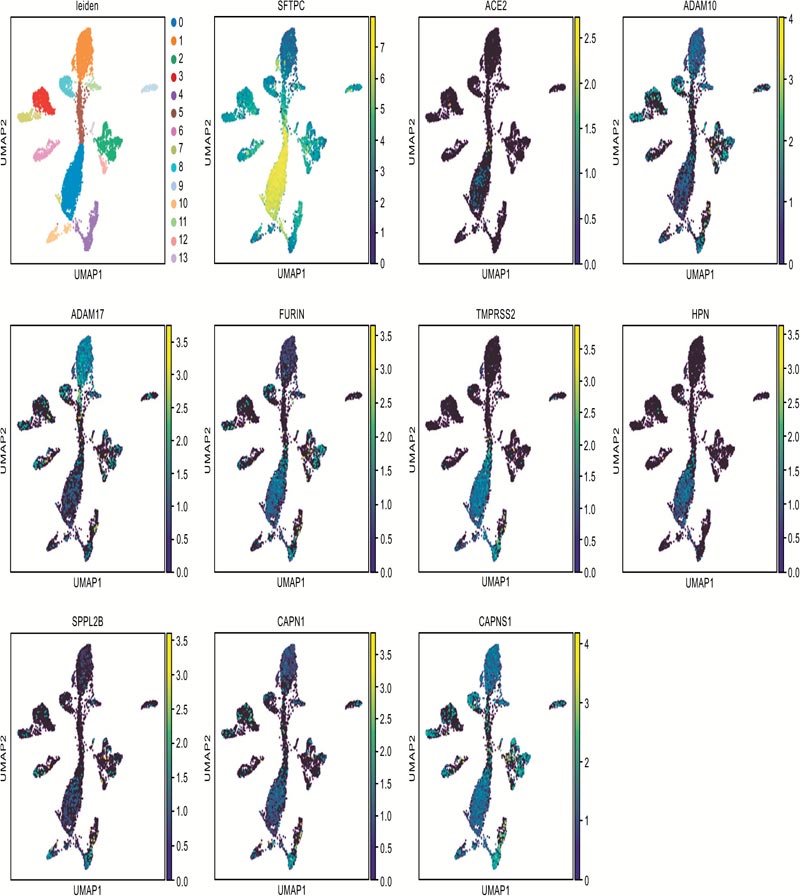
Candidates of membrane endopeptidase selected from lung.
Fig. (1C) illustrates the co-expressed details with ACE2 of each gene. ADAM10 and ADAM17 were reported to cleave the ACE2 to make it shed from cell surface [14, 15]. The cleaving enzyme activity of SPPL2B is similar to FURIN [26]. FURIN is a key factor for cleaving envelope glycoprotein of human immunodeficiency virus type-1 (HIV-1) [27]. In addi- tion, FURIN could proteolytically activate virulent avian influ- enza viruses by cutting their hemagglutinin [28]. TMPRSS2 and HPN both belong to the serine protease family containing a type II transmembrane domain. TMPRSS2 was reported to be involved in the viral entry of SARS-CoV, Sendai virus (SeV), human metapneumovirus (HMPV), human parainfluenza viruses (HPIV) and influenza A virus [4, 29-31]. HPN was reported to be contributed to activate H3N2 influenza A and influenza B Viruses in murine airway [32]. The products of gene CAPN1 and CAPNS1 formed micromolar-calpain (m-calpain). The m-calpain was regarded as an important factor in the replication of SARS-CoV [33]. Hence, these candidates were enrolled in the following research to detect their expression in the target organs.
3.2. The Putative Furin Cleavage Site in the Spike Protein of SARS-CoV-2
As mentioned above, six genes of our candidates, except FURIN and SPPL2B, have been reported to be related to the host cell entry of SARS-CoV. Millet et al. identified the S protein of MERS-CoV could be cleaved and activated by furin at the S1/S2 boundary, which is essential for the MERS-CoV infection [17]. Thus, we wonder whether FURIN would contribute to the viral entry of SARS-CoV-2. To verify this hypothesis, we analyzed the spike protein sequence of SARS-CoV-2. The results indicated that the residue 682-685 cont- ained the minimal furin cleavage motif R-X-X-R [34]. This region was highly similar to the S1/S2 cleavage site of SARS-CoV (Fig. 2A). The results of secondary structure prediction showed this putative furin cleavage site located in the random coil region, which is the preferred target of endopeptidases (Fig. 2B). Hence, we speculated that FURIN might proteo- lytically activate the membrane fusion activity of SARS-CoV-2 through cleaving the spike protein.

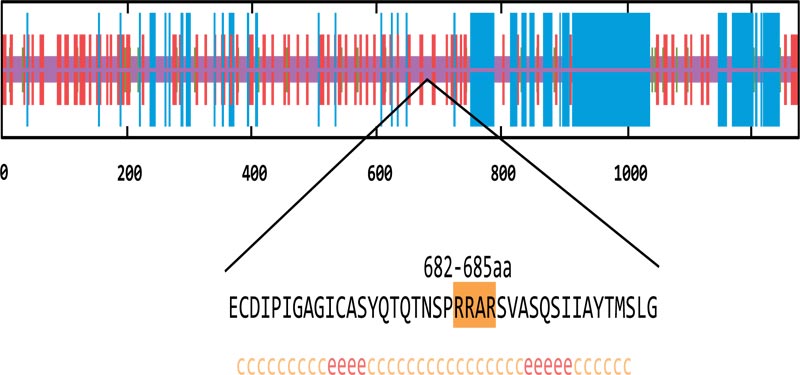
Putative furin cleavage site.
3.3. The Expression of Candidate Endopeptidases in Target Organs
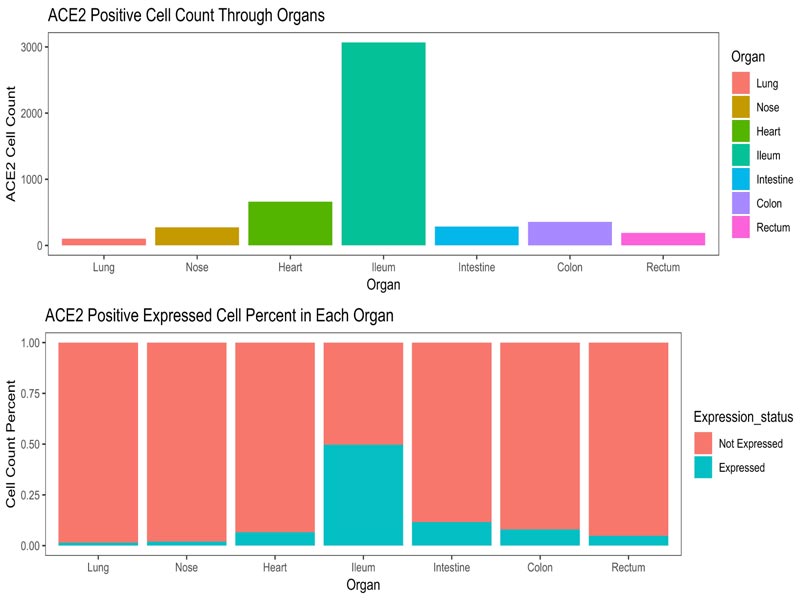
The epithelial cells (EPCAM positive) of nasal brushing samples and nasal turbinate, the cardiomyocytes (MYH6 posi- tive) and endothelial cells (PECAM1 positive) of heart, and the epithelial cells (EPCAM positive) of enteric canal were enrolled into our study. The distribution of ACE2 positive cells in target organs is illustrated in Fig. (3A). The corresponding details about cell counting and percentage were shown in Fig. (3B). As shown in Fig. (3C), the expression of ACE2, TMPRSS2 and CAPNS1 in the lung were abundant, and the FURIN and HPN were moderately expressed. The expression of ACE2 and FURIN in the nose was slightly fewer than those in the lung. TMPRSS2 expression in the nose was significantly less than that in the lung. These results indicate that SARS-CoV-2 could utilize ACE2 and TMPRSS2 to infect airway epithelia, which is similar to the infection mechanism of SARS-CoV. Interestingly, the expressions of CAPN1 and CAPNS1 in the nose were significantly more abundant than that in the lung, indicating that the viruses would replicate rapidly once they could enter the nasal epithelial cells and SARS-CoV-2 might infect nasal epithelia more easily than SARS-CoV.
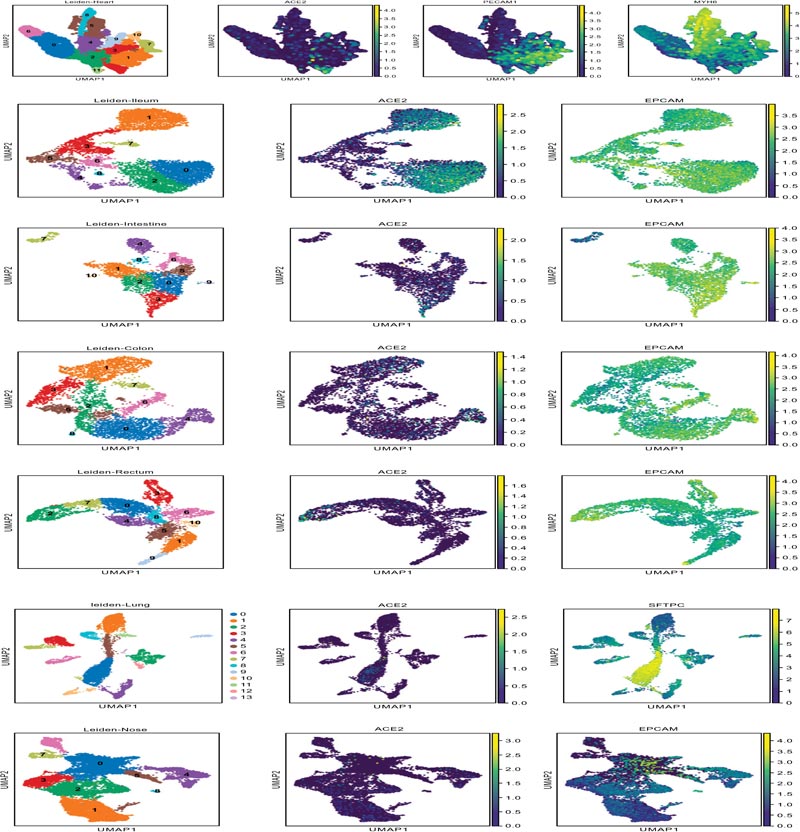

The expression of selected endopeptidases in the target organs.
TMPRSS2, a well-known endopeptidase capable of cleav- ing S protein, was not detected in the cardiac tissue. Additionally, SARS-CoV is unlikely to utilize furin to cleave S protein. Thus, we speculated the lack of endopeptidase-mediated direct virus-cell fusion may partially explain why SARS-CoV rarely causes cardiac damage, despite of the abundant ACE2 expression in cardiomyocytes. Notably, the FURIN expression in cardiac tissue was moderate, which was similar to that in the nose. This suggests SARS-CoV-2 may be able to infect cardiomyocytes and cause cardiac damage, which may contribute to the specific clinical manifestations of COVID-19 patients.
In addition, the expression of ADAM10 and ADAM17 of epithelial cells in enteric canal were abundant. These results suggested that the actual amount of membrane ACE2 would be limited by shedding, despite their ACE2 expression was abun- dant in the normal enterocytes. This finding may be responsible for the relatively milder gastrointestinal discomforts of COVID-19 patients when compared with their respiratory symptoms [3].

The signature score about viral entry of target organs.
3.4. Global Analysis of the Susceptibility of Targeted Organs
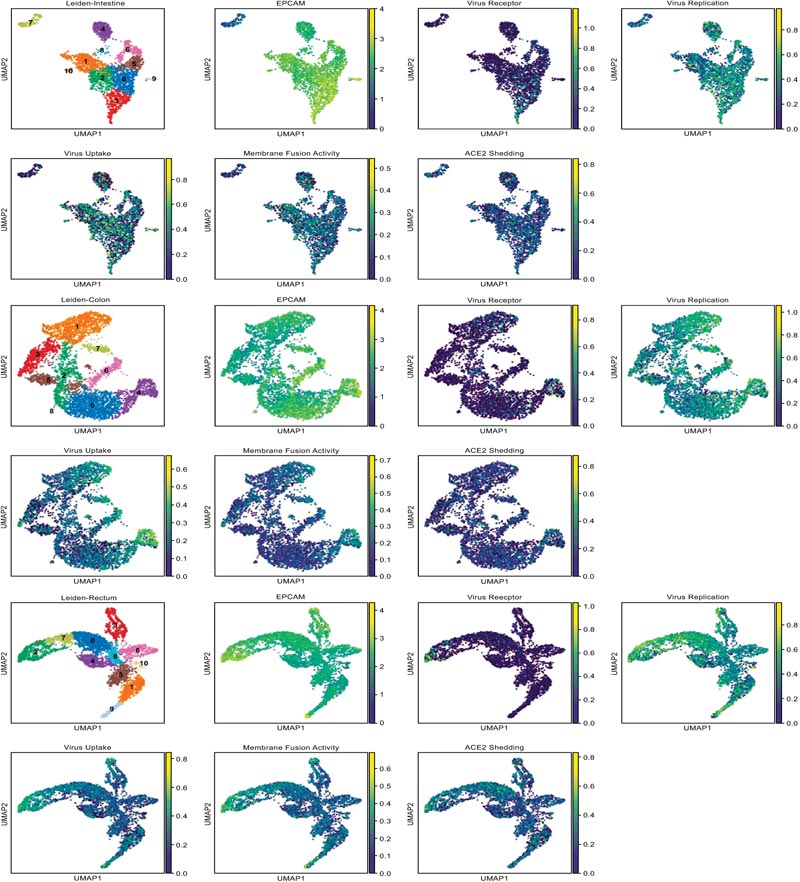
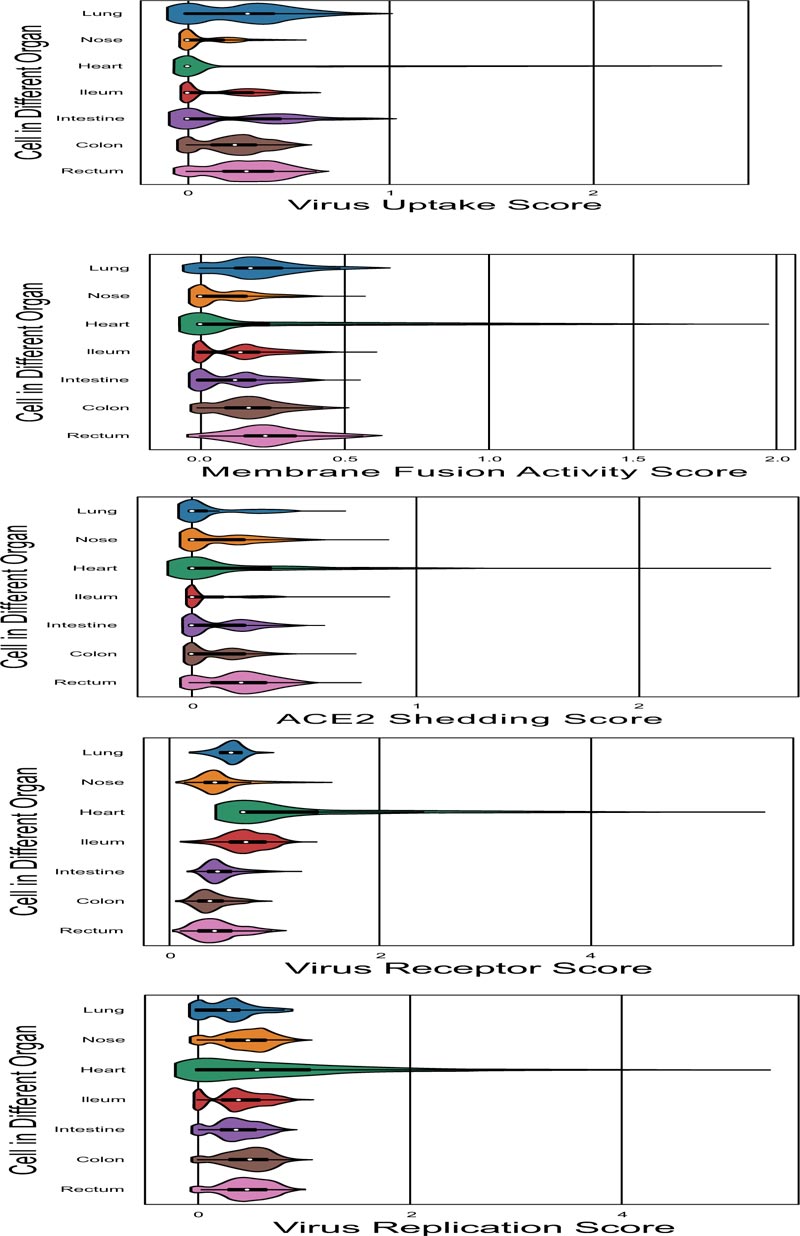
In order to comprehensively assess the efficiency of viral entry to the target organs, we developed a method to calculate the signature score, which could be used as an assessment of each organ cell ability on virus receptor, ACE2 shedding, membrane fusion activity, virus uptake and virus replication function. Gene sets which used to define each function were shown in Fig. (1B). The distribution details of marker genes and function score in target organs were showed in the UMAP scatter plot (Fig. 4A) to illustrate the different function status of cells in a certain organ, and violin plot of signature score about viral entry is shown in Fig. (4B) to illustrate the different function status among the target organs. All of the differences in signature score among target organs were statistically significant, which were calculated through Kruskal-Wallis H test (Fig 4C).
4. DISCUSSION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a lineage B coronavirus, causing the worldwide outbreak of Corona Virus Disease 2019 (COVID-19) [35]. Besides the common respiratory system symptoms, many COVID-19 patients experience gastrointestinal symptoms, car- diac and renal injury and some may even be asymptomatic [3]. The diversity of clinical manifestations challenges the diagnosis of patients and hinders the individualized treatment. Therefore, studies on mechanisms of cell entry could help us comprehensively understand the pathogenesis of SARS-CoV-2 in affected tissues.
The cell surface receptor ACE2 has been proven to facilitate SARS-CoV entry into host cells, and recent studies reveal the crucial role of ACE2 in receptor-mediated SARS-CoV-2 infection as well [36]. What's even more remarkable is that the expression of endopeptidases may provide a 100-fold higher efficient infection pathway than receptor-mediated endocytosis does [37]. In this study, we explored the expre- ssion of screened endopeptidases co-expressed with ACE2 in various targeted cells using the online single-cell sequence datasets. Besides ACE2, the abundant expression of furin and other endopeptidases (such as TMPRSS2, CAPN1 and CAPN- S1) in the nasal epithelia provided prerequisites for SARS-CoV-2 infection and rapid replication in the upper respiratory tract. These findings are in accordance with the recent report of SARS-CoV-2 load in upper respiratory specimens (nasal and throat swabs) [1], suggesting its augmented transmission and requirement for different isolation strategies from SARS-CoV.
Notably, we confirmed a potential furin cleavage site at the S1/S2 interface of SARS-CoV-2 by bioinformatics, which is absent in the S protein of SARS-CoV. This difference between the S protein of SARS-CoV and SARS-CoV-2 was also reported in another recent study from China [38]. Hence, we speculated the presence of furin during the course of SARS-CoV-2 infection may result in some clinical manifestations distinct from SARS-CoV infection. In the case of cardiac tissue, ACE2 expression was detected in the cardiomyocytes, providing the prerequisites for SARS-CoV and SARS-CoV-2 infection. Correspondingly, acute cardiac injury has also been reported in SARS-CoV-2-infected patients, whereas rarely found after SARS-CoV infection [3]. We suppose the furin-mediated cardiomyocyte entry may explain the cardiac damage in some SARS-CoV-2 patients. Following up and evaluation of the cardiac function may be necessary in the SARS-CoV-2-infected patients, especially the elderly patients. Whether the treatment of furin inhibitors could benefit these SARS-CoV-2-infected patients deserves further researches.
Another major point of this study is the global analysis of the susceptibility of potential targeted organs to SARS-CoV-2 infection. Since CoVs commonly infect respiratory and gastro- intestinal systems [39], we take the enteric tissue as an example in the current study. Abundant ACE2 expression was detected in enterocytes from the colon, intestine and rectum, corres- ponding to the enteric symptoms of SARS-CoV-2-infected patients. Meanwhile, the ACE2 sheddase, ADAM 10 and ADAM 17, were also high expressed in the enteric canals, among which the expression of ADAMs was relatively low in the ileum. These findings suggest the actual membrane ACE2 in the ileum epithelia may be higher than that in other enteric canals because of the lower ACE2 shedding rate. Moreover, considering the virus receptor expression, membrane fusion activity, virus uptake and virus replication, the signature score of the ileum was much higher than that in the colon or rectum. Thus, we speculate that the ileum might be more susceptible to the SARS-CoV-2 gastrointestinal infection. Consistent with the recent study [12], this study calls for clinicians to pay attention to the fecal-oral transmission and the enteric damage, especially the ileal abnormalities in COVID-19 patients.
CONCLUSION
In conclusion, our results highlight the potential role of endopeptidase, such as the ACE2 sheddase and furin, in the entry of SARS-CoV-2 into targeted cells. Additionally, we provided a novel method to assess the sensitivities of different targeted organs to SARS-CoV-2 infection, which may benefit future clinical care.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable
HUMAN AND ANIMAL RIGHTS
No humans and animals were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data supporting the findings of the article is available in the Gene Expression Omnibus at [https://www.ncbi. nlm.nih.gov/geo/].
FUNDING
This work was supported by the Key Scientific and Technological Projects of Guangdong Province (2016B0- 90918040, 2017B020230004); the National Natural Science Foundation of China (81970222).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
This work was supported by the Key Scientific and Technological Projects of Guangdong Province (the National Natural Science Foundation of China.


