Multifaceted Disease Diagnosis: Leveraging Transfer Learning with Deep Convolutional Neural Networks on Chest X-Rays for COVID-19, Pneumonia, and Tuberculosis
Abstract
Introduction
The three prevalent yet detrimental respiratory conditions, namely COVID-19, pneumonia, and tuberculosis, exhibit overlapping symptoms, making their differentiation challenging. However, their treatments are significantly divergent. Early detection emerges as a critical common factor for the effective management of these diseases. The pivotal initial step necessitates precise identification to initiate prompt prognosis. However, because of the lack of availability of experts in general and the inadequacy of the medical system on the whole, the problem of early detection is becoming highly concerning and, worst of all, time-consuming.
Objective
This research aimed to address this problem by examining and contrasting various deep Convolutional Neural Network (CNN) models that can accurately identify these illnesses, thereby assisting in their early detection.
Methods
4 pre-trained CNN architectures have been used in this work, namely EfficientNet-B0, VGG-16, InceptionNet, and ResNet-50, which have been implemented on the input dataset. Firstly, the data were collected and pre-processed, and then model training and testing were performed for all 4 pre-trained models specified above.
Results
After fine-tuning the models and evaluating the test metrics on the test dataset, the highest accuracy was observed for ResNet-50 and EfficientNet models, i.e., ~95%. Also, the precision and recall for both were very similar (approximately greater than 92%), indicating accurate and good-quality results.
Conclusion
In this work, a transfer learning system has been employed utilizing several pre-trained CNN architectures. Our findings have indicated that this system can effectively analyze X-ray images to diagnose COVID-19, pneumonia, and tuberculosis.
1. INTRODUCTION
The COVID-19 outbreak has emphasized the critical necessity for precise and effective diagnostic methods. To address this issue, this work has utilized a transfer learning method employing deep CNN to analyze chest X-rays for the identification of COVID-19, pneumonia, and tuberculosis. This approach has shown promising results in accurately detecting these diseases; implementing such a diagnostic approach could make a notable difference in enhancing patient outcomes and alleviating the strain on healthcare systems around the globe. Furthermore, this research study has the potential to pave the way for future developments in diagnostic tools for infectious diseases and could revolutionize the way medical professionals approach the identification and management of respiratory disorders. Additional research is required to authenticate the efficacy of this methodology on larger datasets and to ensure that it can be implemented in a cost-effective and accessible manner, particularly in low-resource settings where these diseases are most prevalent.
Overall, this research paper puts forth an important step in the fight against early detection of these three lung diseases and has the potential to significantly improve the accuracy and efficiency of diagnostic tools, which would ultimately result in improved patient outcomes and alleviate the strain on healthcare systems globally. Policymakers and healthcare professionals must prioritize investments in research and development of innovative diagnostic tools, such as the transfer learning approach proposed in this research paper, to become better equipped to tackle future pandemics and improve global health outcomes. Furthermore, it is important to ensure that these diagnostic tools are attainable and affordable for all individuals, irrespective of their economic or regional status.
This research paper may prove to be valuable in providing revolutionary breakthroughs in the field of diagnostic imaging and tools for respiratory illnesses, highlighting the potential of deep-learning approaches to revolutionize medical diagnosis and treatment. It is hoped that this research work will inspire further collaboration and innovation in the field, ultimately leading to improved global health outcomes and a better quality of life for individuals around the world. Investments in research and development of innovative diagnostic tools must be prioritized, especially when dealing with emerging infectious diseases and other worldwide health concerns, to be prepared for future crises and effectively address the ongoing health needs of individuals and communities worldwide. By doing so, more resilient healthcare systems can be built that are better equipped to respond to emerging health threats and promote the well-being of all individuals, regardless of their background or circumstances.
COVID-19, pneumonia, and tuberculosis are very similar diseases, affecting the respiratory system (majorly the lungs) of human beings. While all three diseases need to be treated differently, in the case of all three, early detection is the key, and expedited treatment can help greatly. In order to treat these diseases, the diagnosis or detection needs to be accurate as these are similar diseases, and an incorrect diagnosis can even lead to death because of negligence. For doing so, several barriers present in the current healthcare systems of most countries, especially the developing ones, need to be highlighted first.
First and foremost, as mentioned previously, the diseases are similar, and their accurate detection is difficult at times due to the amount of correlation present within the diseases. Another issue is the lack of specialist doctors, especially in developing and underdeveloped countries with huge populations. Especially in Southeast Asian countries, like India, Nepal, Vietnam, and Bangladesh, the number of physicians per 1000 population is less than 1, which depicts how busy and engaged the healthcare system is in these countries. Another issue faced is the very long process that takes place during the whole detection phase. The process flow is given below.
As can be seen in Fig. (1), taking multiple appointments, visiting the X-ray clinics multiple times to get the test done, and then getting the reports, all are time-consuming processes and can take a few days to multiple weeks.
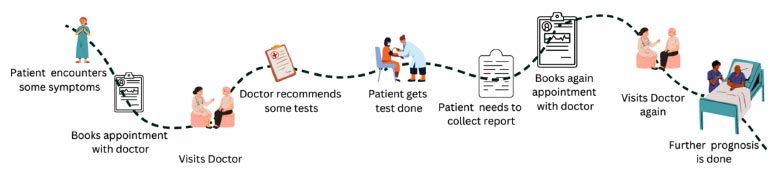
User journey map before the development of the website proposed in this work.
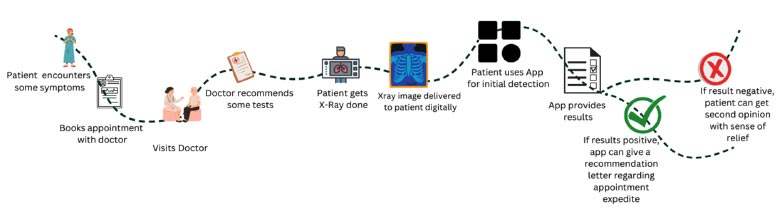
User journey map after the development of the website.
To resolve this problem, a better option could be to use artificial intelligence and build a system that can reduce time and increase convenience for patients and doctors. The revised process flow, being convenient and a bit more effective, is illustrated in Fig. (2).
The main contribution of the work is as follows:
- A model has been developed that is able to classify three lung diseases, namely COVID-19, tuberculosis, and pneumonia, with the help of the transfer learning approach.
- Using Chest X-ray images, the model can predict the presence or absence of the above three diseases.
- Four transfer learning approaches have been applied to the model, namely, EfficientNet, ResNet-50, VGG16, and Inception Net.
- Based on the experimental performance and analysis done, ResNet-50 has been found to provide the most efficient results with 95% accuracy and 97% precision and recall, being the best among all.
2. RELATED WORK
In line with current developments, several techniques are being used to detect COVID-19, pneumonia, and tuberculosis.
Tuberculosis, a global health concern, is caused by Mycobacterium tuberculosis. The number of cases of tuberculosis is increasing by millions each year. Currently, six nations report the highest number of cases, and the common factor linking the nations is sizable regions with low economic growth. There are few doctors and medical services in these places [1, 2]. The proposed system was assessed using three distinct classification techniques: nearest neighbour, logistic regression, and support vector machine [3]. Some medical facilities in nations with few resources and dense populations lack the essential or modern equipment to conduct Chest X-ray (CXR) screening [4]. The patients are requested to have a CXR taken, and there exists a significant chance that they have Tuberculosis (TB). Consequently, the traditional TB diagnosis process may take a long time [5]. X-ray image-based diagnosis is a tedious and time-consuming task, requiring significant manual effort and therefore being prone to human error [6-8]. Though numerous AI-based methods for diagnosing TB now exist, researchers are still working toward achieving the necessary accuracy [9]. Also, another work utilized ResNet, Xception, Inception-ResNet-V2, MobileNet, and EfficientNet, employing the same approach [9-13]. Qualified radiologists conduct manual analysis and CXR detection, which is a time-consuming and difficult task [14]. Additionally, the low-resource nations, particularly the rural areas, lack qualified radiologists [15].
The COVID-19 pandemic has severely impacted global health and general welfare. A crucial measure in the battle against COVID-19 is the successful identification of infected individuals with radiology examination utilizing chest radiography as a vital screening technique. The most serious illness caused by COVID-19, such as pneumonia, impacts the lungs [16]. The testing procedures, such as Reverse Transcription Polymerase Chain Reaction (RT-PCR), are time-consuming and sometimes even fatal [17-19]. Other diagnostic methods for COVID-19 include examining clinical symptoms, investigating epidemio- logical background, and analyzing positive radiographic images (such as CT or CXR). These methods can cause the process to become lengthy and are less accurate [18, 20]. Different CNN models used are unable to work accurately with images having tilted or faulty corners. Many research studies have advocated using transfer learning and deep CNN for diagnostic purposes [21-25]. Many methods, such as DeTraC, have been suggested in which frameworks, like VGG16, are used [26].
Furthermore, pneumonia is a disease that impacts either or both lungs and causes irritation of the air sacs contained within. These sacs get filled with pus or fluid, leading to symptoms, such as difficulty in breathing, fever, chills, and a productive cough that generates pus or phlegm. A study [27] showed how Bayesian optimization may be used to detect and diagnose pneumonia using CNN architecture estimates. A Deep Learning (DL)-based strategy has also been put forth to identify and categorize pneumonia from chest X-ray images [28-31]. The three pre-existing architectures, ResNet50, InceptionV3, and InceptionResNetV2, have been utilized in a transfer learning technique to extract both temporal and spatial features [32]. A thorough analysis of deep learning applications on X-ray images of the chest has made it abundantly obvious that the quantity of the dataset affects the system's effectiveness. Therefore, a combination of various data augmentation techniques can lead to better results [33-35].
One thing that most studies have lacked is the presence of a one-stop solution to classify these diseases. This means that most studies have only tried to detect either COVID-19, pneumonia, or tuberculosis individually; however, a few studies have aimed to detect two or more diseases together.
This paper has focused on the analysis and study of the transfer learning approach, applying a few pre-trained CNN models on real-world chest X-ray datasets to predict and classify three lung diseases, i.e., COVID-19, pneumonia, and tuberculosis.
3. MATERIAL AND METHODOLOGY
3.1. Dataset
We have combined publicly available datasets for all three diseases, consisting of a total of 4300 images, namely, 700 tuberculosis X-ray images obtained from Kaggle by Tawisfur Rahman, 1200 COVID-19 X-ray images taken from Kaggle by Prashant Patel, 1200 pneumonia X-ray images (including bacterial and viral pneumonia) obtained from Kaggle by Paul Mooney, and 1200 normal X-ray images taken from GitHub. These X-ray images had different size ranges. It, therefore, became important to standardize the image size of all images obtained from different data sources. We standardized the images to 224 pixels x 224 pixels. Furthermore, standardizing the image colour of all images obtained from different data sources was also an important aspect of data preparation. One-hot encoding on labels also played a key role in this study. All datasets were of the same classes, corresponding to 4 classes in total. Some examples of X-ray images indicating COVID-19 infection are presented in Fig. (3). Fig. (4) displays several examples of X-ray images indicating positive cases of tuberculosis. Fig. (5) showcases multiple instances of X-ray images indicating positive cases of pneumonia. Finally, Fig. (6) illustrates a range of normal X-ray images where no evidence of the three lung diseases is present.
3.2. CNN and DL
Deep learning-based medical image analysis has drawn a lot of interest recently because of its potential to help radiologists diagnose medical diseases more effectively and accurately. Convolutional Neural Networks (CNNs) in particular have become an effective tool for picture segmentation and classification in the medical field, including the diagnosis of various lung illnesses.
COVID-19-positive CXR images.
TB-positive CXR images.
Pneumonia-positive CXR images.
Normal CXR images.
This paper aimed to present a thorough evaluation of current research, utilizing deep learning and CNNs for the detection of lung diseases in medical images, including pneumonia, tuberculosis, lung cancer, and COVID-19. Fig. (7) illustrates the proposed methodology. This study has also examined the datasets, architectures, and performance metrics used earlier and discussed their potential uses for medical image analysis, and proposed future directions for this technology.
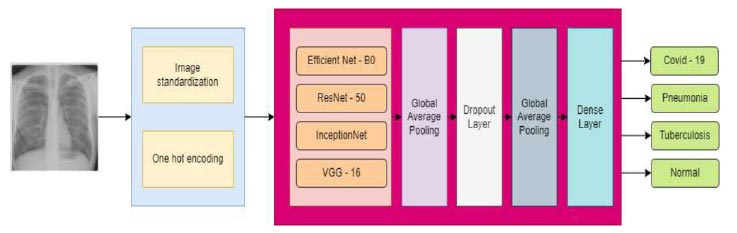
Proposed methodology for multi-disease detection.
According to our findings, CNNs and deep learning offer a lot of potential to help radiologists identify and categorize lung disorders. However, before they may be used as common diagnostic tools, further research must be done to confirm the precision and generalizability of these models in clinical settings. These findings may be encouraging and point to a bright future in the application of deep learning and CNNs in the analysis of medical images and the identification of illnesses. It might further lead to better patient outcomes and lower healthcare costs.
Overall, this work emphasizes the significance of research and development in deep learning and medical image analysis. It shows the potential to fundamentally change medical diagnoses and significantly enhance patient care.
3.3. Transfer Learning
The method of Transfer Learning (TL) involves fine-tuning a model with pre-trained weights from a larger dataset, and is widely used in deep learning on a smaller or more specific dataset. The method of transfer learning is especially beneficial in the field of medical image analysis due to the scarcity of large, annotated datasets. In the case of CXR images, TL has been shown to be effective in predicting COVID-19, pneumonia, and Tuberculosis (TB) using a variety of deep learning models.
To use TL for CXR images in the prediction of COVID-19, pneumonia, and TB, the following steps must be taken:
3.3.1. Obtain a Pre-trained Model
There are several pre-trained models available for chest X-ray image analysis, such as VGG, ResNet50, and Inception. These models have been trained on bigger datasets, and therefore, their weights may be used as a starting point for training on a smaller medical image dataset.
3.3.2. Fine-tune the Model
The pre-trained model can then be fine-tuned on a smaller dataset of CXR images, labeled for COVID-19, pneumonia, and TB. This involves modifying the final layers of the model to match the classes in the dataset and training the model on the new dataset while keeping the weights from pre-trained models fixed.
3.3.3. Evaluate the Model
One can evaluate the performance of the fine-tuned model on a validation set by analyzing metrics, like precision, recall, accuracy, and F1-score.
3.3.4. Test the Model
After the evaluation of the model, it can be tested on a distinct test set to measure its practical performance.
In conclusion, transfer learning is a powerful technique for CXR image analysis in COVID-19, pneumonia, and TB prediction. It allows deep learning models to leverage pre-existing knowledge from large datasets and fine-tune smaller medical image datasets to achieve high accuracy and performance.
3.4. Training and Model Building
In our investigation, we meticulously assessed the performance of pre-trained Convolutional Neural Networks (CNNs) by adapting them to our dataset individually. To accomplish this, we employed four prominent pre-trained CNN architectures: EfficientNetB0, InceptionV3, ResNet50, and VGG16. Prior to model training, thorough data preparation was performed, involving the conversion of images into numerical values and carrying out one-hot encoding, along with standardizing the image size. Notably, we ensured standardization across the Chest X-ray (CXR) images to mitigate any potential issues arising from color and size variations, thus maintaining consistency in the dataset. The ample size of the dataset provided sufficient data for robust model training, facilitating reliable results.
For fine-tuning each pre-trained CNN architecture, we primarily focused on the convolutional part of their respective architectures. Following this, we augmented each network with a global average pooling layer and a dense layer, which were appended to the final convolutional layer. Subsequently, a classification layer utilizing a dense layer with SoftMax activation was added to enable multi-class classification. Fine-tuning of all networks was conducted using Stochastic Gradient Descent (SGD) optimization with the Adam optimizer, employing a batch size of 32 over 5 epochs. Categorical cross-entropy served as the loss function for all models, and hyperparameters were fine-tuned based on performance metrics evaluated on the validation set.
During the initial stage of data preparation, particular attention was given to resizing the images and organizing them into distinct folders based on the required input size of each model, ensuring compatibility with their respective architectures. A comparative analysis of the performance of the four pre-trained CNN models is presented in Table 1, wherein consistent initialization and learning rate policies have been employed during training to facilitate fair evaluation and comparison.
| Loss | Categorical cross-entropy |
|---|---|
| Optimizer | Adam |
| Metrics | Accuracy |
| Monitor | Validation accuracy |
| Epochs | 5 |
| Batch Size | 32 |
| Input image size | 224 x 224 x 3 |
| Weights | ImageNet |
By meticulously configuring and fine-tuning each pre-trained CNN architecture, we aimed to optimize their performance for the task of lung pathology detection in X-ray images. This rigorous approach ensures that the selected models are adeptly tailored to the dataset, maximizing their efficacy in accurately identifying and classifying lung diseases.
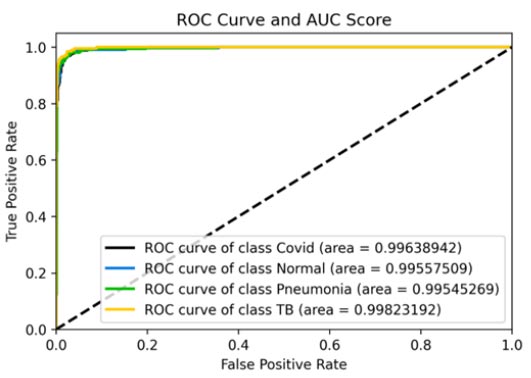
ROC curve and AUC score.
Precision-recall curve.
Confusion matrix.
Classification report.
For all 4 models, the following hyperparameters were used:
3.4.1. Evaluation Metrics
Four distinct pre-trained CNN models were independently fine-tuned to our dataset, and subsequently, their predictions were utilized on the test set. The dataset was partitioned into training, validation, and testing sets with proportions of 70%, 9%, and 21%, respectively. The proposed method was evaluated using standard classification metrics, including accuracy, precision, recall, F1 score, and the area under the Receiver Operating Characteristic (ROC) curve known as AUC. The F1-score is a statistical measure that computes the harmonic mean of precision and recall. On the other hand, the AUC or area under the curve is a performance metric that assesses the ability of a classification model to distinguish between classes by evaluating various threshold settings.
4. RESULTS
Four pre-trained CNN models, including Effici-entNetB0, InceptionV3, ResNet-50, and VGG16, were fine-tuned to our dataset containing CXR images of COVID-19, normal, TB, and pneumonia cases. The aim was to transfer the pre-existing knowledge from these models to aid our task, which had limited training data. The deep learning process was implemented using Keras software and the Python programming language. The accuracy and loss function values for the training sets during the fine-tuning of various pre-trained CNN models are depicted in the following figure. Diagnostic accuracy was computed using the test set after five training iterations. The outcomes of transfer learning post-employment of different architectures are presented in Table 1, including standard classification metrics, such as accuracy, precision, recall, F1 score, and AUC.
4.1. EffecientNetB0 Model
Fig. (8a and b) and b show the ROC curve obtained for the EffecientNetB0 model. The accuracy obtained for the model was roughly 95%. The confusion matrix presented in Fig. (8c) can be used to effectively infer further results regarding the same. The ROC-AUC curves provided in Fig. (8d) can also be used to gain further inferences about this model in its entirety. The precision-recall curves for all classes have also been suggestive of the effectiveness of this model. The precision (94%) for this model was also relatively high, suggesting it to be a very good model for the dataset. The recall (97%) and F1-score (96%) were also the highest for this model, which again indicated the supremacy of this model.
4.2. ResNet-50 Model
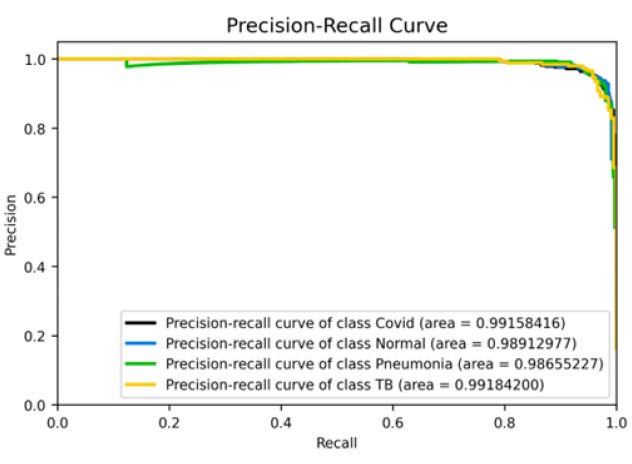
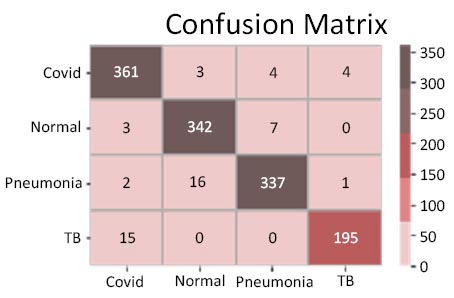
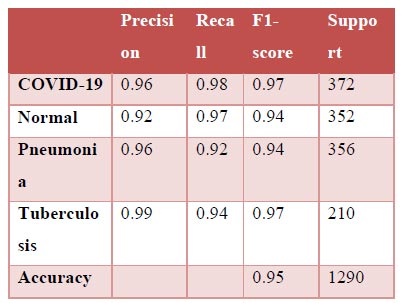
The results shown in Fig. (9) pertain to the ResNet50 model, which achieved an accuracy of approximately 96%. By analyzing the confusion matrix, we could obtain additional insights into the model's performance. Additionally, ROC-AUC curves can provide valuable information about the model's effectiveness. The precision-recall curves for all categories suggested the high efficiency of the model. The model's precision rate of 96% was notably high, indicating it to perform well on the dataset. Furthermore, the recall rate (97%) for this model and F1-score (96%) were among the highest, further underscoring its quality.
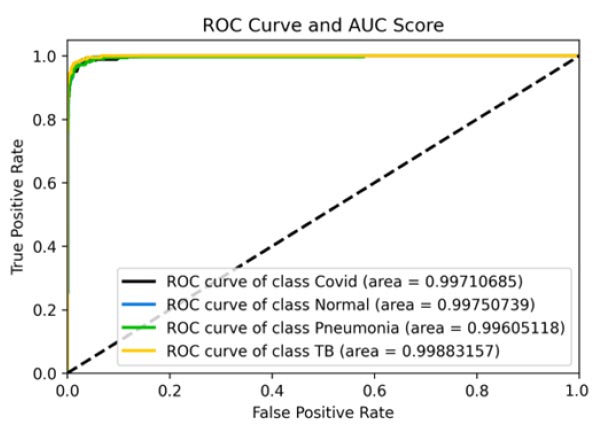
ROC curve and AUC score.
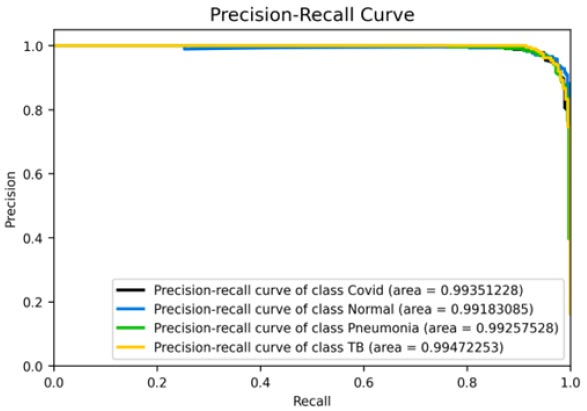
Precision-recall curve.
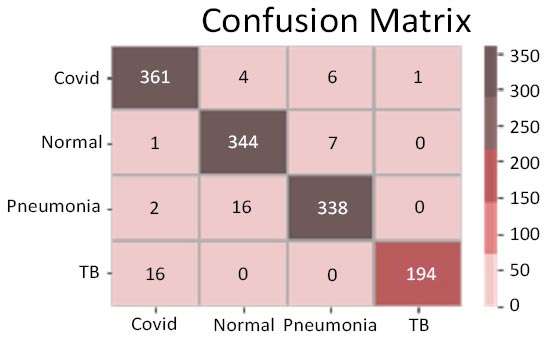
Confusion matrix.
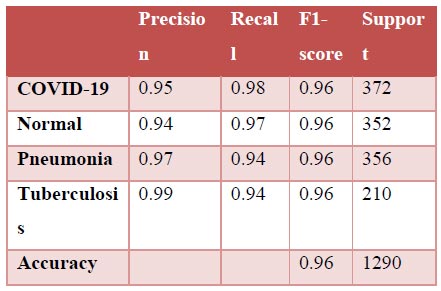
Classification report.
4.3. VGG-16 Model
Fig. (10a-d) show the results of the VGG16 model that achieved an accuracy score of approximately 93%, which has been found to be significantly good, but not as good as the previous two models. Analysing the confusion matrix can provide further insights into the model's performance. The analyses have suggested this model to rank average among all 4 models. Moreover, the ROC-AUC curves showed the model to be relatively good but lesser than the previous two models. The precision-recall curves for all categories suggested the model to be highly efficient, but that it can be fine-tuned to provide better results and improved scores. Its precision rate of 96% has been found to be among the highest of the 4 models, indicating it to perform remarkably well on the given dataset. Furthermore, the model's recall rate and F1-score, both at 91% and 94%, respectively, have not been found to be as high as the previous two models; nonetheless, they have highlighted its quality. Overall, the results have suggested the VGG16 model to be a strong performer and that it can be considered reliable for the given task in case the previous two models face difficulties, but it would further need changes to be the frontrunner if selected as a primary predictor model.
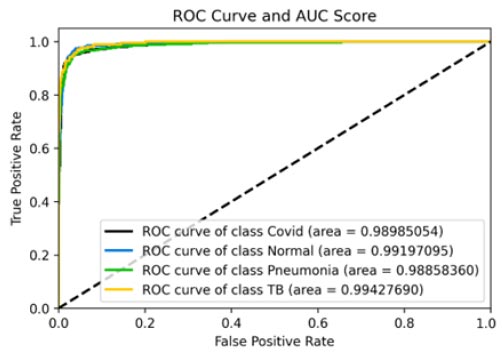
ROC curve and AUC score.
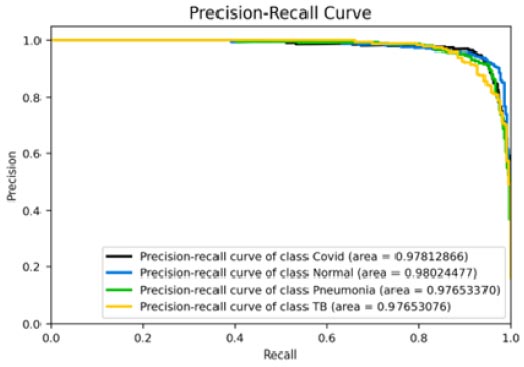
Precision-recall curve.
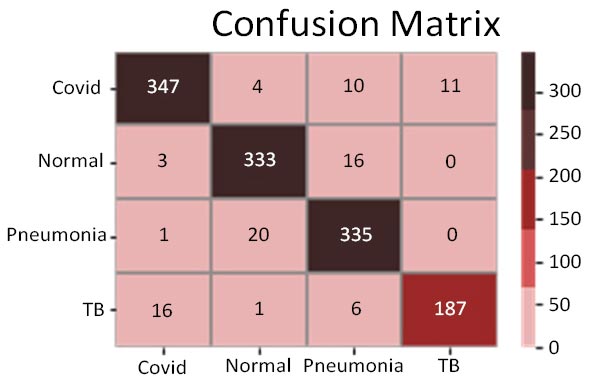
Confusion matrix.
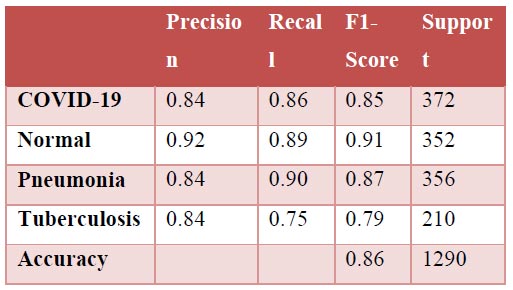
Classification report.
4.4. InceptionNet Model
Fig. (11a-d) demonstrate the results of the InceptionV3 model, which attained an accuracy of roughly 86%, being phenomenal by all means, but still not comparable with the other three models discussed earlier in terms of accuracy. Further insights into the model's performance can be derived by analysing the confusion matrix. Additionally, the ROC-AUC curves have shown the model to not perform bad. However, it has not been found to be effective enough or reliable when compared to the three models discussed before. The precision-recall curves for all classes have suggested the model to be effective enough, but not significantly reliable in comparison. However, keeping in mind its direct relation to the medical field, this model may not suffice in terms of reliability. With a precision rate of 81%, the model performed well on the dataset, but it can be fine-tuned in the future to produce better results. Moreover, the model's recall rate (90%) and F1-score (85%) have been found to be the lowest of all, rendering it to be not appealing enough. In some scenarios, this particular model might not be as convenient as the previous three models.
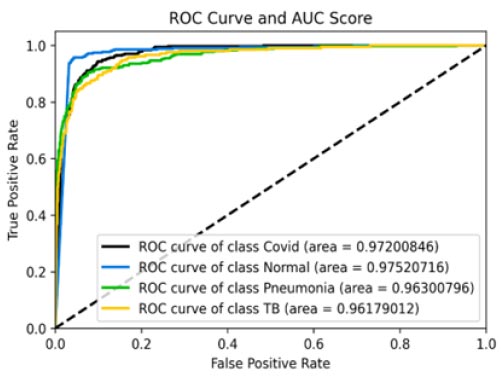
ROC curve and AUC score.
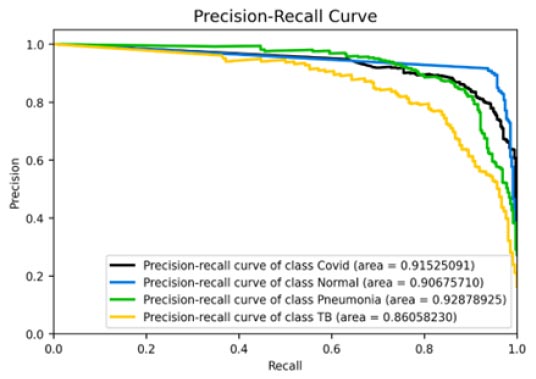
Precision-recall curve.
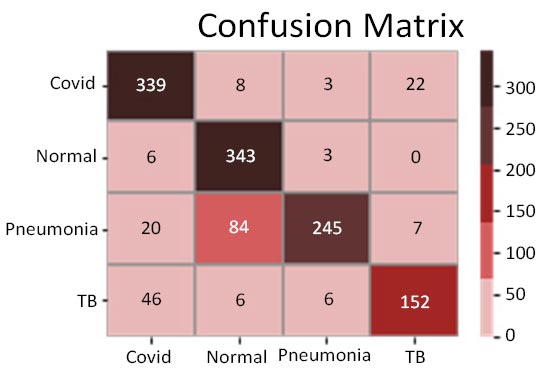
Confusion matrix.
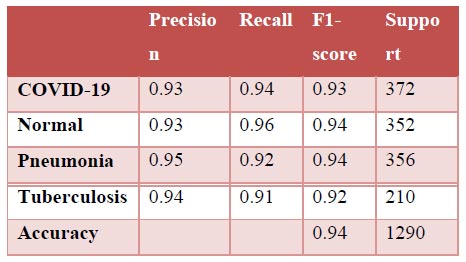
Classification report.
| - | Model | |||
|---|---|---|---|---|
| Metrics | EfficientNet | ResNet50 | InceptionNet | VGG16 |
| Precision | 0.94 ± 0.03 | 0.96 ± 0.03 | 0.81 ± 0.08 | 0.96 ± 0.08 |
| Recall | 0.97 ± 0.06 | 0.97 ± 0.04 | 0.90 ± 0.30 | 0.91 ± 0.04 |
| F1-score | 0.96 ± 0.02 | 0.96 ± 0.01 | 0.85 ± 0.13 | 0.94 ± 0.03 |
| Accuracy | 0.96 | 0.95 | 0.86 | 0.93 |
| AUC (COVID-19) | 0.99638942 | 0.99710685 | 0.97200846 | 0.98985054 |
| AUC (Normal) | 0.99557509 | 0.99750739 | 0.97520716 | 0.99197095 |
| AUC (Pneumonia) | 0.99545269 | 0.99605118 | 0.96300796 | 0.9885836 |
| AUC (TB) | 0.99823192 | 0.99883157 | 0.96179012 | 0.9942769 |
4.5. Performance Review
The experimental findings presented in Table 2 illustrate the performance evaluation metrics of four distinct Convolutional Neural Network (CNN) models, EfficientNet, ResNet50, InceptionNet, and VGG16, in the context of early detection of lung diseases, encompassing COVID-19, pneumonia, Tuberculosis (TB), and normal cases. Precision metrics have revealed EfficientNet and ResNet50 to excel in minimizing false positives across all classes, while InceptionNet has exhibited comparatively lower precision, particularly for COVID-19 and pneumonia. Robust recall values of EfficientNet and ResNet50 have underscored their ability to effectively capture positive instances with minimal false negatives. The F1-score analysis has emphasized the balanced performance of EfficientNet and ResNet50, with InceptionNet showcasing lower scores primarily attributed to reduced precision. Notably, AUC values have highlighted the superior discrimination capabilities of EfficientNet and ResNet50 across all disease categories. These findings collectively suggest EfficientNet and ResNet50 to be promising candidates for accurate and reliable early detection of lung diseases, with potential implications for enhancing diagnostic precision in clinical settings.
5. DISCUSSION
In recent years, the integration of Artificial Intelligence (AI) and deep learning techniques has revolutionized medical diagnostics, particularly in the field of radiology. In this work, we have introduced a robust Convolutional Neural Network (CNN) framework tailored for the autonomous identification of lung diseases from X-ray images. Leveraging the power of state-of-the-art pre-trained CNN models, EfficientNet, ResNet50, VGG16, and InceptionNet, our study has focused on the automated detection of three significant lung pathologies, including COVID-19, pneumonia, and Tuberculosis (TB).
The selection of pre-trained CNN models has been driven by their proven performance across a diverse range of datasets from publicly available sources. This approach has ensured a strong foundation for our framework, enabling efficient feature extraction and learning representations from the X-ray images. To tailor these models specifically for the task at hand, we have employed fine-tuning techniques, allowing the networks to adapt to the intricacies of lung pathology detection. The process has involved training the networks on a substantial quantity of data, being crucial for achieving optimal performance.
In our evaluation, the results have highlighted the superior performance of the EfficientNet model compared to other pre-trained CNN models. With an accuracy of 0.96, precision of 0.94, and recall of 0.97 in classifying X-ray images, the EfficientNet model has emerged as the frontrunner, demonstrating its efficacy in accurately identifying lung pathologies, which underscores its potential as a robust approach to diagnosing lung diseases from chest X-ray images.
To enhance usability and accessibility, we have developed a user interface to streamline the process of inputting X-ray images for predictions regarding lung health. The system has incorporated checkpoints to dynamically assess and switch to a more accurate model, ensuring optimal model utilization for each image. Additionally, we have addressed the challenge of limited training data by proposing an ensemble technique based on majority voting among diverse deep transfer learning outputs. This approach has mitigated the issue of dataset size and enhanced the reliability of our predictions.
Looking ahead, our future plans involve expanding the dataset to encompass a broader range of X-ray images, including those from diverse demographics and clinical settings. We aim to further validate the effectiveness and reliability of our proposed approach through extensive testing and evaluation. Ethical considerations, interpretability, and human supervision are critical aspects that we would continue to prioritize, ensuring the ethical and responsible deployment of AI in medical diagnostics.
In conclusion, the comprehensive approach presented in this investigation holds promise as a valuable tool in the identification of lung diseases in X-ray images. However, ongoing scrutiny and validation are imperative to affirm its precision and reliability in real-world medical scenarios. By advancing the intersection of AI and medical imaging, we aim to contribute to improved healthcare outcomes and patient care.
While our study has demonstrated promising results in the automated identification of lung diseases from X-ray images using pre-trained CNN models, it is essential to acknowledge several limitations that could impact the generalizability and practical applicability of our findings. Firstly, our analysis has been contingent upon the quality and representativeness of the dataset utilized for model training and evaluation. Despite efforts to curate a diverse dataset, inherent biases may have existed, potentially skewing the performance of the models towards certain demographics or disease manifestations. To address these biases, efforts are required to collect and annotate data from diverse patient populations and clinical settings, ensuring the robustness and inclusivity of the model's predictions.
Furthermore, while our models have demonstrated high accuracy and performance metrics in controlled experimental settings, their efficacy in real-world clinical scenarios may be influenced by various factors. These include differences in image acquisition techniques, variations in disease presentation, and the presence of confounding factors not accounted for during model training. Thus, further validation studies in clinical settings are imperative to assess the models' performance under real-world conditions, including their ability to integrate seamlessly into existing healthcare workflows without disrupting clinical practices.
Additionally, the successful integration of AI-based diagnostic tools into clinical practice hinges on considerations beyond model performance alone. Practical challenges, such as regulatory compliance, data privacy concerns, and workflow integration, must be carefully addressed to ensure seamless adoption by healthcare providers. Moreover, effective communication strategies are essential to facilitate collaboration between AI systems and human clinicians, fostering trust and transparency in decision-making processes.
In light of these limitations, future work should prioritize efforts to enhance the robustness and generalizability of AI models for lung disease detection. This includes ongoing data collection efforts to diversify and expand the training dataset, as well as the development of interpretable AI techniques to elucidate the rationale behind model predictions. Moreover, collaboration with healthcare stakeholders is essential to iteratively refine AI models and ensure their alignment with clinical needs and practices. Ultimately, by addressing these limitations and challenges, AI-based diagnostic tools have the potential to augment and enhance clinical decision-making, ultimately improving patient outcomes in the diagnosis and management of lung diseases [36-40].
CONCLUSION
This research study’s findings indicate the feasibility of diagnosing COVID-19, pneumonia, and TB through chest X-rays utilizing the transfer learning approach with deep Convolutional Neural Networks (CNNs). By employing a chest X-ray dataset, the study has applied transfer learning using four distinct pre-trained CNN architectures: EfficientNetB0, VGG-16, InceptionV3, and ResNet-50. The methodology has encompassed data collection, pre-processing, and comprehensive model training and testing across all four pre-trained models. Post-fine-tuning and evaluation of test metrics on the test dataset, the study has observed ResNet-50 and EfficientNet models to achieve the highest accuracy, both nearing 96%, with EfficientNet exhibiting a slightly superior performance. Comparable precision and recall values for both models have underscored the accuracy and quality of the obtained results. The findings suggest that leveraging deep learning with transfer learning methodologies holds promise in developing automated systems for the early identification and diagnosis of specific illnesses, especially in resource-limited settings. The integration of chest X-rays with deep learning strategies could contribute to the creation of more efficient and precise diagnostic tools for a spectrum of medical conditions. Furthermore, the potential of deep learning in addressing diverse medical imaging tasks for disease detection warrants exploration in future research endeavours. In summary, this study has provided valuable insights into the application of deep learning techniques for illness diagnosis, indicating significant potential for advancing healthcare outcomes in the future.
AUTHORS’ CONTRIBUTION
It is hereby acknowledged that all authors have accepted responsibility for the manuscript's content and consented to itssubmission. They have meticulously reviewed all results and unanimously approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| AI | = Artificial intelligence |
| CNN | = Convolutional neural network |
| TB | = Tuberculosis |
AVAILABILITY OF DATA AND MATERIALS
The data and supportive information are available within the article.


