The Common Ancestor of Deinococcus Species was Rod-Shaped
Abstract
Background:
The genus Deinococcus consists of species in rod-shape (Bacilli) and spherical shape (Cocci).
Objective:
In this study, we aimed to determine whether the common ancestor of Deinococcus species was rod-shaped or spherical.
Methods:
We compared the homologs of the proteins related to the rod-shape in bacteria (MreB, MreC, MreD, MrdA, RodA, and RodZ) in various Deinococcus species and Thermus thermophilus.
Results:
The phylogenetic trees based on each protein and the homologs reflected the evolutionary relationships of the species, indicating that the Horizontal transfer of the genes did not occur during the Deinococcus evolution.
Conclusion:
The ancestor of the genus Deinococcus was rod-shaped, and the spherical forms appeared when the rod-shaped formation system was lost during evolution and diversification within the genus.
1. INTRODUCTION
In 1981, the genus Deinococcus was defined as cocci [1]. The radiation-resistant spherical bacterium Deinococcus radiodurans has a unique lipid composition, i.e., glycolipids and glycophospholipids have been identified, but phosphatidylethanolamine and phosphatidylglycerol, which are common in other bacteria, are absent [2-4]. This unique character is observed in this bacterium [5]. In 1987, Deinobacter grandis was reported as a radiation-resistant rod-shaped bacterium [6]. In 1997, D. grandis was transferred to the genus Deinococcus based on 16S rDNA sequence comparison [7]. At present, the genus Deinococcus consists of both bacilli and cocci species. Evolutionarily, the order Deinococcales is closely related to the order Thermales [8]. These two orders constitute the phylum Deinococcus-Thermus. The purpose of this study was to elucidate whether the common ancestor of the Deinococcus species was spherical or rod-shaped on the basis of molecular evolutionary analyses. Although the last common ancestor of bacteria has been thought to be rod-shaped [9], the rod-shaped related proteins (MreB, MreC, MreD, MrdA, RodA, and RodZ) have been recently reported [10-13]. In spherical bacterium Staphylococcus aureus, MreC and MreD are not essential for cell viability and do not affect cell morphology [14]. If the genes coding for those six proteins had been inherited during the evolution of Deinococcus species, their common ancestor would be rod-shaped. If those genes had been acquired by horizontal transfer, the common ancestor would probably be spherical.
2. MATERIALS AND METHODS
In this study, we used the complete genome sequences of eleven species, and the almost complete genome sequences of three species (D. ficus, D. grandis, and D. hopiensis) of Deinococcus (Table 1). Homologs of Thermus thermophilus were used as out-groups. Orthologous protein sequence comparison is a powerful tool that enables a more accurate phylogenetic evaluation than that based on 16S and/or 23S rRNA sequence comparison [15]. Most orthologous proteins are ribosomal proteins [16]. Thus, in order to understand the evolutionary relationships among Deinococcus species, a phylogenetic tree was constructed using their ribosomal protein sequences. The amino acid sequences of the ribosomal proteins- S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, S12, S13, S14, S15, S16, S17, S18, S19, and S20 of the small subunit, and L1, L2, L3, L4, L5, L6, L7/L12, L9, L10, L11, L13, L14, L15, L16, L17, L18, L19, L20, L21, L22, L23, L24, L25, L27, L28, L29, L30, L31, L32, L33, L34, L35, and L36 of the large subunit, were used for the tree construction. We identified proteins, homologous to each of the six proteins, which are related to rod-shape in bacteria, namely MreB, MreC, MreD, MrdA, RodA, and RodZ, in each of the fifteen bacterial species considered in the study. Homologous proteins were selected on the basis of BLASTP search in NCBI. Multiple sequence alignment using MUSCLE [17] and phylogenetic tree construction in Maximum Likelihood (ML) method using MEGA software, version 6 were performed [18]. The Le_Gascuel_2008 (LG) model [19] was selected as the best model for each ML analysis. The nearest neighbor interchange was used as the heuristic approach for the ML method for inferring the phylogenetic tree. The γ-distribution rate was considered, and the number of discrete γ-categories was five. Bootstrap analysis was performed with 1000 replicates.
| Organism | Shape | Genome GC% | Assembly level (NCBI Accession Number) | MreB Homolog | MreC Homolog | MreD Homolog | MrdA(PBP2) Homolog | RodA Homolog | RodZ Homolog |
|---|---|---|---|---|---|---|---|---|---|
| Deinococcus actinosclerus | rod/spherical | 70.6 | complete (CP013910.1, CP029774.1) | present | present | present | present | present | present |
| Deinococcus deserti | rod | 63.0 | complete (CP001114.1) | present | present | present | present | present | present |
| Deinococcus ficus | rod | 69.9 | 6 scaffolds (ATTJ00000000.1) | present | present | present | present | present | present |
| Deinococcus geothermalis | spherical | 66.6 | complete (CP000359.1) | lack | lack | lack | lack | lack | lack |
| Deinococcus gobiensis | spherical | 69.2 | complete (CP002191.1) | lack | lack | lack | lack | lack | lack |
| Deinococcus grandis | rod | 69.9 | 7 scaffolds (BCMS00000000.1) | present | present | lack | present | present | present |
| Deinococcus hopiensis | spherical | 64.9 | 11 scaffolds (FWWU00000000.1) | lack | lack | lack | lack | lack | lack |
| Deinococcus maricopensis | rod | 69.8 | complete (CP002454.1) | present | present | lack | present | present | present |
| Deinococcus peraridilitoris | short rod/spherical | 63.7 | complete (CP003382.1) | present | present | present | present | present | present |
| Deinococcus proteolyticus | spherical | 65.7 | complete (CP002536.1) | lack | lack | lack | lack | lack | lack |
| Deinococcus puniceus | spherical | 62.6 | complete (CP011387.1) | present | present | present | present | present | present |
| Deinococcus radiodurans | spherical | 66.7 | complete (AE000513.1, AE001825.1, CP015081.1, CP015082.1) | lack | lack | lack | lack | lack | lack |
| Deinococcus soli | short rod | 70.2 | complete (CP011389.1) | present | present | lack | present | present | present |
| Deinococcus swuensis | spherical | 67.4 | complete (CP010028.1) | present | present | present | lack | present | lack |
3. RESULTS AND DISCUSSION
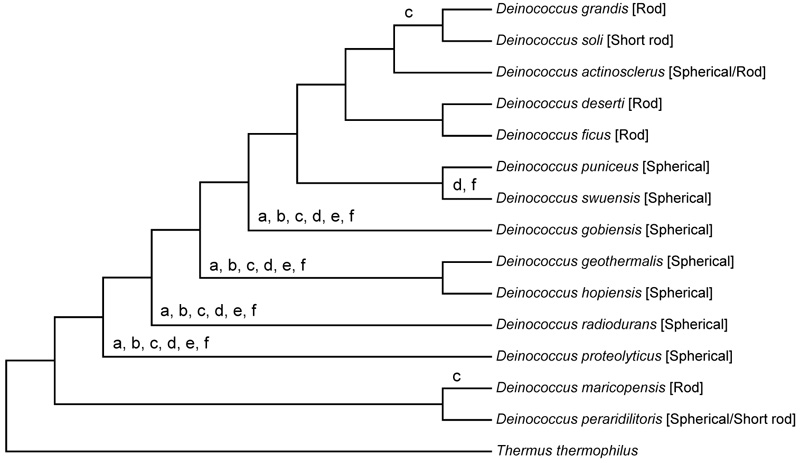
The ribosomal protein sequence comparison produced more accurate phylogenetic trees than the rRNA sequence comparison (Fig. 1). The phylogenetic tree based on the ribosomal protein sequences showed that neither bacilli nor cocci formed a monophyletic lineage; both the spherical and rod-shaped species were scattered in the tree (Fig. 1).

Phylogenetic tree based on ribosomal proteins: The evolutionary history was inferred using the maximum likelihood method based on the Le_Gascuel_2008 model [19]. The tree with the highest log likelihood (-63291.1582) is shown. The percentage of trees in which the associated taxa were clustered together is shown next to the branches. Initial tree(s) for heuristic search were obtained by applying the neighbor-joining method to a matrix of pair wise distances estimated using a JTT model [20]. A discrete γ-distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.4545)). The tree was drawn in scale, with branch lengths measured in the number of substitutions per site. The analysis involved 15 amino acid sequences. All positions containing gaps and missing data were eliminated. There were totally 6904 positions in the final set of data. Evolutionary analyses were conducted using MEGA 6 software [18].
Phylogenetic tree based on rRNAs: The evolutionary history was inferred using the maximum likelihood method based on the Tamura-Nei model [21]. The tree with the highest log likelihood (-19723.9210) is shown. The percentage of trees in which the associated taxa were clustered together is shown next to the branches. Initial tree(s) for heuristic search were obtained by applying the neighbor-joining method to a matrix of pair wise distances estimated using the maximum composite likelihood (MCL) approach. A discrete γ-distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.1283)). The tree was drawn in scale, with branch lengths measured in the number of substitutions per site. The analysis involved 15 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were totally 4160 positions in the final set of data. Evolutionary analyses were conducted using MEGA 6 software [18].
The results revealed that most of the rod-shape related genes were not only distributed in bacilli, but also in cocci. For example, the spherical D. puniceus was shown to have all of the six rod-shape related genes (Table 1). It is uncertain whether all of the six genes are expressed and function in D. puniceus. On the other hand, the rod-shaped D. grandis and D. maricopensis lacked mreD homologs (Table 1). In addition, D. actinosclerus and D. peraridilitoris, which can become bacilli or cocci, showed to have all six rod-shape related genes (Table 1).
Among eleven complete genomes of Deinococcus analyzed, D. actinosclerus, D. deserti, D. peraridilitoris, and D. puniceus had all the six rod-shape related genes (Table 1). Homologs of three genes among the six, mreC, mreD, and mrdA, formed a conserved gene cluster, which were found conserved in T. thermophilus also (Fig. 2). Although mrdA and rodA were found to be clustered in Escherichia coli [22, 23], mrdA homolog was not clustered with rodA homolog, but with mreC and mreD homologs in Deinococcus and T. thermophilus (Fig. 2). Homologs of the other three genes mreB, rodA, and rodZ were scattered in the genome of all the species (Fig. 2). Although mreB, mreC, and mreD were found clustered in E. coli [24], mreB homolog was not clustered with mreC and mreD homologs in Deinococcus and T. thermophilus (Fig. 2). These results indicate that horizontal transfer of the gene cluster did not occur.

Phylogenetic relationships inferred on the basis of rod-shape related genes showed that phylogenetic clusters were conserved. D. actinosclerus, D. grandis, and D. soli were found to be clustered when analyzed for their mreB, mreC, rodA, and rodZ homologs; D. deserti and D. ficus were clustered for their mreB, mreC, mreD, and mrdA homologs (Fig. 3). In addition, all phylogenetic trees deduced in this way indicated that D. maricopensis and D. peraridilitoris had diverged prior to the separation of other Deinococcus species (Fig. 3), which is in agreement with the evolution of Deinococcus species, elucidated from the ribosomal protein sequence comparison (Fig. 1).
The evolutionary history was inferred using the maximum likelihood method based on the Le_Gascuel_2008 model [19]. Phylogenetic trees based on rod-shape related proteins: The tree with the highest log likelihood (-2103.0678 in MreB, -2842.1661 in MreC, -1771.9295 in MreD, -6392.3576 in MrdA, -3330.3680 in RodA, and -3760.3540 in RodZ) is shown. The percentage of trees in which the associated taxa were clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pair wise distances estimated using a JTT model. A discrete γ-distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.5099 in MreB, 0.9195 in MreC, 1.1260 in MreD, 0.6919 in MrdA, 0.9724 in RodA, and 1.0505 in RodZ)). The tree is drawn in scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. There were totally 345, 240, 152, 550, 342, and 270 positions of MreB, MreC, MreD, MrdA, RodA, and RodZ respectively, in the final set of data. Evolutionary analyses were conducted using MEGA 6 software [18].

CONCLUSION
Our results showed that each of the rod-shaped related genes had been inherited in most of rod-shaped species of Deinococcus during the evolution of Deinococcus species. Thus, the common ancestor of Deinococcus species was rod-shaped. Major gene loss had occurred four times during the evolution of Deinococcus with respect to the rod-shape related genes, which led to the generation of cocci in the same genus (Fig. 4).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICTS OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors thank Dr. Issay Narumi for valuable comments.


