All published articles of this journal are available on ScienceDirect.
Unsupervised Deep learning-based Feature Fusion Approach for Detection and Analysis of COVID-19 using X-ray and CT Images
Abstract
Aims:
This study investigates an unsupervised deep learning-based feature fusion approach for the detection and analysis of COVID-19 using chest X-ray (CXR) and Computed tomography (CT) images.
Background:
The outbreak of COVID-19 has affected millions of people all around the world and the disease is diagnosed by the reverse transcription-polymerase chain reaction (RT-PCR) test which suffers from a lower viral load, and sampling error, etc. Computed tomography (CT) and chest X-ray (CXR) scans can be examined as most infected people suffer from lungs infection. Both CT and CXR imaging techniques are useful for the COVID-19 diagnosis at an early stage and it is an alternative to the RT-PCR test.
Objective:
The manual diagnosis of CT scans and CXR images are labour-intensive and consumes a lot of time. To handle this situation, many AI-based solutions are researched including deep learning-based detection models, which can be used to help the radiologist to make a better diagnosis. However, the availability of annotated data for COVID-19 detection is limited due to the need for domain expertise and expensive annotation cost. Also, most existing state-of-the-art deep learning-based detection models follow a supervised learning approach. Therefore, in this work, we have explored various unsupervised learning models for COVID-19 detection which does not need a labelled dataset.
Methods:
In this work, we propose an unsupervised deep learning-based COVID-19 detection approach that incorporates the feature fusion method for performance enhancement. Four different sets of experiments are run on both CT and CXR scan datasets where convolutional autoencoders, pre-trained CNNs, hybrid, and PCA-based models are used for feature extraction and K-means and GMM techniques are used for clustering.
Results:
The maximum accuracy of 84% is achieved by the model Autoencoder3-ResNet50 (GMM) on the CT dataset and for the CXR dataset, both Autoencoder1-VGG16 (KMeans and GMM) models achieved 70% accuracy.
Conclusion:
Our proposed deep unsupervised learning, feature fusion-based COVID-19 detection approach achieved promising results on both datasets. It also outperforms four well-known existing unsupervised approaches.
1. INTRODUCTION
Coronavirus disease 2019, often referred to as COVID-19, is a deadly disease that affected millions of people all around the world. It is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV 2). In the past, the world has witnessed other viruses like MERS, SARS, etc [1]. that only lasted for a few days or months [2]. Unlike other cases, COVID-19 spread all around the world within four months and now it has become a global pandemic, affecting more than 3 million people and causing more than a quarter-million deaths by the month of May 2022. Unfortunately, there is no confirmed COVID-19 vaccine released yet for mass public usage.
At present, the standard diagnosis of COVID-19 is made using the reverse transcription-polymerase chain reaction (RT-PCR) test that uses swabs from the nasopharynx area of the throat, but it suffers from disadvantages like lower viral load, sampling error, etc [3]. Besides, due to the shortage of RT-PCR test kits, many test cases cannot be run. Most of the people who were affected by COVID-19 suffer from pneumonia and radiographic imaging techniques such as chest X-rays (CXR) and computed tomography (CT) scans may be useful for the diagnosis at an early stage and it is an alternative to RT-PCR test. The initial patient presentation using CT scan screening shows that it outperforms the RT-PCR approach in terms of sensitivity [4] and it also confirmed COVID-19 for some of the cases which are diagnosed incorrectly as negative by the RT-PCR method [3].
Apart from CT scans, CXR scans are also commonly used for diagnosing pneumonia throughout the world as they are cheap and fast [5, 6]. However, the manual diagnosis of COVID-19 using both methods needs domain expertise and experience. Fortunately, deep learning-based automatic COVID-19 detection models based on CT scans and CXR images are explored by many researchers since deep learning models have the ability to extract and learn from optimal features automatically with very little human intervention to make the accurate decision [7-10]. These AI-powered COVID-19 detection models can be used for accurate detection as well as to aid the radiologist to improve the time and accuracy of their diagnosis [11].
Almost all of the state-of-the-art deep learning-based COVID-19 classification models are supervised learning models [12, 13]. Supervised learning-based models require annotated datasets in order to learn during the training process. Collection and manual annotation of CT scans and CXR images are expensive and time-consuming and require expert knowledge. Also, the biggest challenge in the classification of medical images is the limited availability of annotated images. Therefore, in this work, we have explored various unsupervised learning models for COVID-19 detection, which do not need a labelled dataset.
We have used convolutional autoencoder networks, PCA-based models, pretrained CNN models and Hybrid feature fusion based models for feature extraction and KNN and GMM for clustering. We have trained our models for both CT and CXR scan datasets and we have analysed their performance. The main contributions of our models are:
- We have employed many unsupervised methods such as convolutional autoencoders, PCA-based models, pretrained CNN ImageNet architectures for feature extraction, along with clustering methods such as Kmeans and GMM for unsupervised learning based COVID-19 detection using CT and CXR scan datasets.
- We have also proposed a hybrid unsupervised deep learning approach based on the feature fusion technique for COVID-19 detection.
- The training, validation, and testing of various unsupervised deep learning-based models are shown on big CT and CXR datasets.
- We have analysed the performance of our models using the standard metrics and visualized the features using t-SNE.
- The performance of the existing three unsupervised methods is shown for COVID-19 detection using both CT and CXR scan datasets.
- The performance of the proposed approach based on an unsupervised deep learning-based feature fusion approach is compared with the existing four well-known unsupervised methods.
The remaining sections of this paper are organized as follows; Section 2 contains the related work. Section 3 provides the description of the dataset. Section 4 includes the methodology. Section 5 presents the results and analysis. Section 6 includes discussions and future works and Section 7 concludes the work.
2. RELATED WORK
In the past, several research works based on deep learning-based COVID-19 detection models that work using CT and CXR scan datasets have been proposed. Bai et al. [11] propose an EfficientNet-based abnormal chest computed tomography (CT) scan image classification model to distinguish COVID-19 disease from other pneumonia. They performed external validation that is ignored by most studies, and their model achieved good performance when compared to the manual diagnosis by many radiologists. The authors suggested that the diagnostic accuracy of radiologists has improved significantly with the help of their AI model. Ardakani et al. [14] studied the effectiveness of various well-known CNN architectures like ResNet, Xception, etc. for COVID-19 detection. Ten binary models were trained using chest CT images and the ResNet-101 model achieved the best performance (AUC of 0.994) among them. The second-best performance is shown by the Xception model.
Kang et al. [15] proposed an automatic multi-view representation learning-based COVID-19 diagnostic framework that extracts different kinds of features from the CT scans to differentiate COVID-19 pneumonia from community-acquired pneumonia (CAP). The authors suggest that these extracted features can be utilized together to significantly improve the performance of the detection model. The proposed model achieves an accuracy of 95.5%. Similarly, the work [5] by Ouyang et al. proposes an online CNN-based attention network to use the information from the infected region for enhancing the classification of COVID-19 pneumonia from CAP. They have used the dual sampling (size-based and uniform) method to handle imbalanced learning. Their method achieves an AUC of 0.94.
Wang et al. [16] propose a coronavirus detection framework that uses prior attention residual learning techniques to learn better discriminative features for COVID-19 classification. The proposed work uses two 3D ResNet models in which one performs binary pneumonia classification (normal or pneumonia) and the other one takes the attention information regarding lesion regions passed from the prior model to improve the performance of classification of COVID-19 and interstitial lung disease pneumonia images.
Wang et al. [18] proposed a deep learning-based COVID-19 detection and lesion localization framework. Their model takes segmented lung images as input from UNet and predicts the COVID-19 probability. It also performs lesion localization without the need for annotation. Their model achieves an AUC of 0.96 and it takes nearly 2 seconds to handle a CT image. The authors have released their framework for public access. Similar to a study [18], this work [20] proposes an automated deep learning-based framework to differentiate coronavirus cases from CAP and non-pneumonia cases and also to locate the lesion region.
Harmon et al. [19] proposed a deep learning-based multi-modal approach for detecting the COVID-19 probability. Three AI models are trained; the first one is a CNN model that uses segmented chest CT images and the second one is machine learning models that are trained on other clinical information. The third multi-modal network combines both clinical information and features from CT scan images using an MLP network. The joint model has achieved an AUC of 0.92.
Oh et al. [21] proposed a multiple patch-based convolutional neural network model that takes a small amount of segmented CXR images as input and uses a majority voting method to determine the probability of COVID-19 disease with a very less number of trainable parameters. They also proposed a new gradient-weighted class activation map to interpret the results in a meaningful way. In this study [22], the authors studied the performance of 4 popular CNN-based transfer learning architectures such as DenseNet-121, SqueezeNet, ResNet50, and ResNet18 for chest radiograph classification, and some of them have achieved a maximum true positive rate of 98%. They visualized the infected region using heatmaps. They publicly released their dataset, code, and models [23].
Wang et al. [24] proposed a deep CNN model named COVID-Net for the classification of normal, COVID-19, and non-COVID-19 cases using CXR scan images. They also developed a benchmark dataset of almost 14,000 CXR scans that is publicly available. They incorporated model explainability approaches to gain insights into model predictions to aid clinical diagnosis and their model achieved an accuracy of 93.3%. Narin et al. [25] studied the performance of pretrained CNN models for COVID-19 detection on 3 different datasets. Their models are Inception-ResNetV2, InceptionV3, ResNet152, ResNet101, and ResNet50 and unlike the previous works, they have four different classes such as normal, COVID-19, and other pneumonia cases (bacterial & viral). The authors reported that among the tested models, ResNet50 showed superior performance [26].
Apostolopoulos et al. [27] studied the effectiveness of many state-of-the-art CNN-based ImageNet transfer learning models for COVID-19 detection. The pretrained models such as Inception-ResNet-V2, Xception, Inception, MobileNet-V2, and VGG-19 are trained to classify CXR scan images into normal, COVID-19, and other cases of pneumonia (viral & bacterial) classes. Among the studied models, VGG-19 is the best performing one with an accuracy of 98.8% and MobileNet-V2 is the second-best performing model with an accuracy of 97.4%. According to another study by Abbas et al. [28], the performance of such pretrained models for COVID-19 detection can be further enhanced by incorporating the class decomposition layer. In this study, the CXR scan images are classified into normal, COVID-19, and Non-COVID-19 (SARS) categories using a transfer learning-based model named DeTraC [29] which handles the irregularities in the dataset (data imbalance and intensity of inhomogeneous images) by subdividing each class into various sub-classes and assembles them back before the end prediction. The performance of the DeTraC model was compared with models such as SqueezeNet, GoogleNet, ResNet, VGG, and AlexNet and the authors reported that the DeTraC model shows superior performance with a maximum accuracy of 93.1%.
As discussed in the introduction section, our literature review found that there is a lack of research publication related to unsupervised learning approaches to detect COVID-19 using CT and CXR scan images. The work [30] is one of the studies which applies unsupervised clustering for the detection of COVID-19 using CXR images. It employs a self-organizing feature map network to extract optimal features and cluster the unlabelled data. A self-organizing feature map is an iterative approach that requires a good initial approximation to achieve optimal performance. Besides, the performance of the self-organizing feature map was shown on a small dataset. Therefore, due to this lack of publications related to unsupervised learning-based methods and limited availability of annotated data, we have trained many unsupervised learning models and we have studied their performance.
3. DATASET
Two different datasets [31] are used in our work; one is CT scan images and the other one is chest X-ray images which are released by Mendeley [32]. We have used 8,055 and 9,544 CT scans and X-ray images respectively. Detailed statistics of the aforementioned datasets are listed in (Table 1). Each dataset is divided such that 20% of the data is used for testing the models and the rest of the 80% of data is further divided such that 55% of data is used for training the models and the other 25% of data is used for the purpose of validation. The datasets of CT and X-ray are divided into train, valid, and test datasets randomly. The sample images of the CT scan and X-ray dataset are shown in Fig. (1a and 1b) respectively. In each of these images, the first row contains image samples for COVID-19 and the second row contains image samples for Non-COVID-19.

| Dataset | COVID-19 | Non-COVID-19 | Total |
|---|---|---|---|
| CT | 5,427 | 2,628 | 8,055 |
| X-ray | 4,044 | 5,500 | 9,544 |
4. METHODOLOGY
In this work, we have used various unsupervised models for feature extraction and K-means and GMM for clustering the features into COVID-19 or Non-COVID-19. The various unsupervised models are PCA, Convolutional autoencoder, pretrained VGG-16, VGG-19, and ResNet50 models. In this section, the brief details of the aforementioned various unsupervised models and the proposed methodology are included.
4.1.1. Principal Component Analysis-based Models
Principal Component Analysis (PCA) is a well-known unsupervised statistical approach used in machine learning mostly for reducing the dimensionality of the data [37]. Both images from both datasets are passed to the PCA method with a variance parameter of 0.98. The dimensionality of CT scans and X-ray images is reduced from 4,096 to 855 and 4,096 to 788 dimensions, respectively. Fig. (1) shows the original and retained chest X-ray and CT scan images before and after passing the PCA method. The obtained optimal features are clustered into two groups representing COVID-19 and Non-COVID-19 classes using the Kmeans clustering algorithm.
4.1.2. Convolutional Autoencoder Models
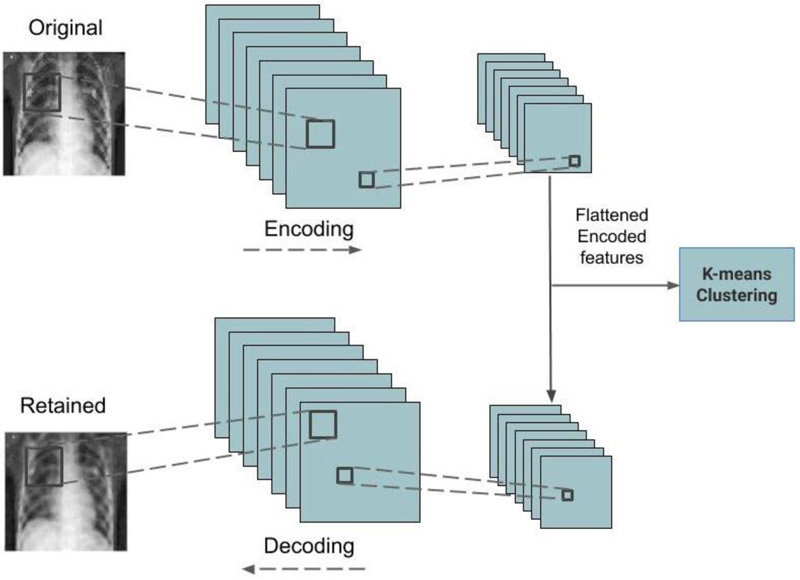
The convolutional autoencoder is an unsupervised learning network that uses convolution operations to reproduce the images that are passed as input in the output layer [37]. The encoder module compresses the images which are reproduced by the decoder module. The compressed features are extracted from the encoder and fed into the Kmeans function for the purpose of clustering them. Fig. (2) represents the architecture of the trained convolutional autoencoder used in this work. Four convolutional autoencoder models are trained for both datasets. Adam method is used as the optimizer function and the mean squared error method is used as the loss function. Drop out regularization is incorporated to handle model overfitting. Table 2 lists the layer type, output dimension, and parameter count of the best performing convolutional autoencoder model for the CT scan image dataset, which has a total of 793,537 trainable parameters. Similar to this, the best performed convolutional autoencoder details for the CXR dataset are shown in Table 3 containing a total of 48,577 trainable parameters.
| Model Layer | Output Shape | #Parameters |
|---|---|---|
| Conv2D | (-, 64, 64, 256) | 7168 |
| MaxPooling2D | (-, 32, 32, 256) | - |
| Conv2D | (-, 32, 32, 128) | 295040 |
| MaxPooling2D | (-, 16, 16, 128) | - |
| Conv2D | (-, 16, 16, 64) | 73792 |
| MaxPooling2D | (-, 8, 8, 64) | - |
| Conv2D | (-, 8, 8, 32) | 18464 |
| MaxPooling2D | (-, 4, 4, 32) | - |
| Conv2D | (-, 4, 4, 32) | 9248 |
| UpSampling2D | (-, 8, 8, 32) | - |
| Conv2D | (-, 8, 8, 64) | 18496 |
| Model Layer | Output Shape | #parameters |
|---|---|---|
| Conv2D | (-, 64, 64, 64) | 1792 |
| MaxPooling2D | (-, 32, 32, 64) | - |
| Conv2D | (-, 32, 32, 32) | 18464 |
| MaxPooling2D | (-, 16, 16, 32) | - |
| Conv2D | (-, 16, 16, 32) | 9248 |
| UpSampling2D | (-, 32, 32, 32) | - |
| Conv2D | (-, 32, 32, 64) | 18496 |
| UpSampling2D | (-, 64, 64, 64) | - |
| Conv2D | (-, 64, 64, 1) | 577 |
4.1.3. Pretrained CNN Models
Pretrained CNN ImageNet models are well-known and commonly used in most computer vision tasks such as image processing, recognition, etc. [37]. and this work uses these transfer learning-based models such as VGG16, VGG19, and ResNet50 for feature extraction. The top or last layer of the network is removed and the remaining layers are frozen after loading the ImageNet weights. Images from both datasets are resized into 64X64x3 dimensions and fed into the models. The outputs features are then flattened, passed to the PCA method for dimensionality reduction, and further Kmeans or GMM methods are applied for clustering them into two separate clusters.
4.1.4. Hybrid Models using Feature Fusion Method
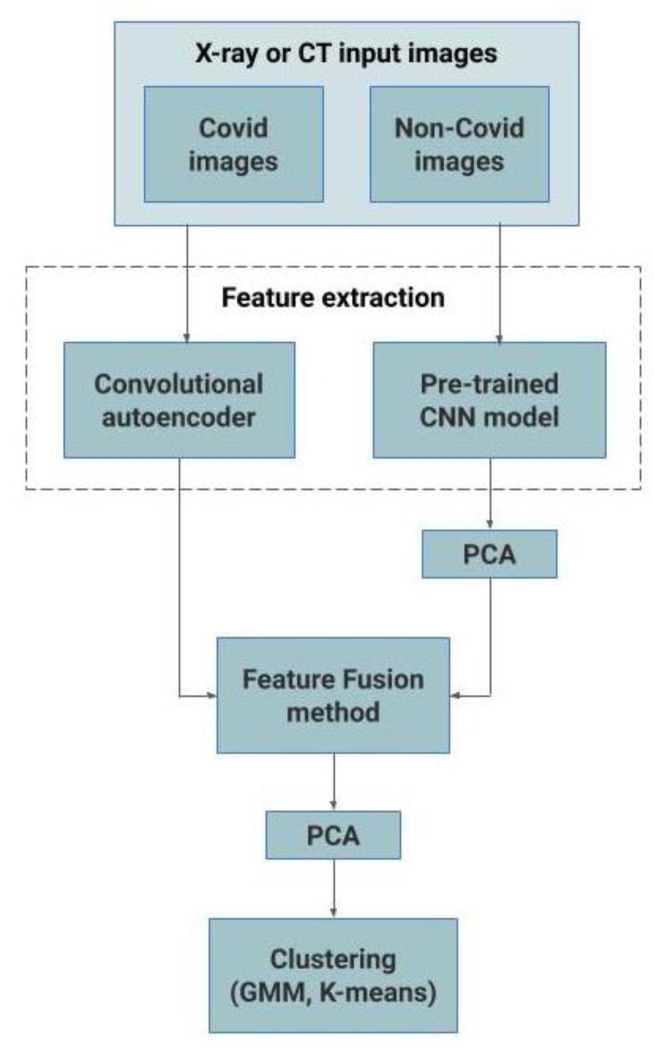
For the performance enhancement, the output of the best performing convolutional autoencoder model and pretrained CNN model are merged using the feature fusion method, and finally, PCA is applied before clustering them using Kmeans or GMM methods. Fig. (3) represents the proposed feature fusion-based approach. For the CT scan dataset, the output of autoencoder3 and ResNet50 models are merged and for the CXR dataset, the output of autoen coder1 and VGG-16 models are merged. This feature fusion-based approach has resulted in significant improvement in performance. The pseudo-code of this approach is given as Algorithm 1.
5. EXPERIMENTAL RESULTS
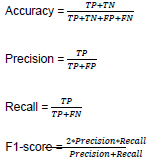
All the models that are used in this work are implemented using Keras and TensorFlow python library and the implementation is publicly available in Github3. Google Colab4is used in this work to run all the experiments. The Google Colab has a Tesla K80 GPU of about 25GB RAM. The performance of all the models is reported using standard metrics such as accuracy, precision, recall, and f1-score which are obtained from the confusion matrix. The confusion matrix is constructed using terms such as true positive (TP), true negative (TN), false negative (FN), and false positive (FP). These terms along with the standard metrics are defined with their equations below:
- TP = Count of instances of COVID-19 samples are predicted accurately as COVID-19.
- FN = Count of instances of COVID-19 samples are incorrectly predicted as COVID-19.
- FP = Count of instances of Non-COVID-19 samples are incorrectly predicted as COVID-19.
- TN = Count of instances of Non-COVID-19 samples are predicted accurately as Non-COVID-19.
 |
In this work, four different sets of experiments are conducted on both CT and CXR scan images. Initially, 4 different convolutional autoencoder models along with Kmeans are used for feature extraction and clustering, respectively. All the models are trained for 100 epochs with a batch size of 64, and the ReLU activation function is used. For all models learning rate is 0.01 and no momentum is used. (Table 4) presents the performance of 4 convolutional autoencoder and PCA-based models for the CT scan dataset. For the CT scan dataset, among the four models, the autoencoder3 model with Kmeans performed better with an accuracy of 67%, and its model layer details are listed in Table 2. For all the Kmeans experiments, n_clusters=8, init='k-means++', n_init=10, max_iter=300, and tol=0.0001 is used.
Algorithm 1 COVID-19 detection using hybrid feature fusion-based approach
Input: A set of input COVID-19 and Non-COVID-19 sample images x1, x2, ..., xn (CT or CXR scans)
Output: A set of labels y1, y2, ..., yn corresponding to each input based on COVID-19 and Non-COVID-19 clusters.
1: Resize and normalize the input images.
//Extracting features f from Convolutional autoencoder (CA) and pretrained CNN (pCNN) models.
2: f1 = CA(normalized input images)
3: f2 = pCNN(normalized input images)
4: f2pca = P CA(f2)
//Merging both the features using feature fusion method
5: f f1 = featurefusion(f1, f2pca)
6: f f1pca = PCA(f f1)
//Clustering (K-Means or GMM) the fused features into COVID-19 and Non-COVID-19 clusters
7: Output labels y =Clustering(f f1pca)
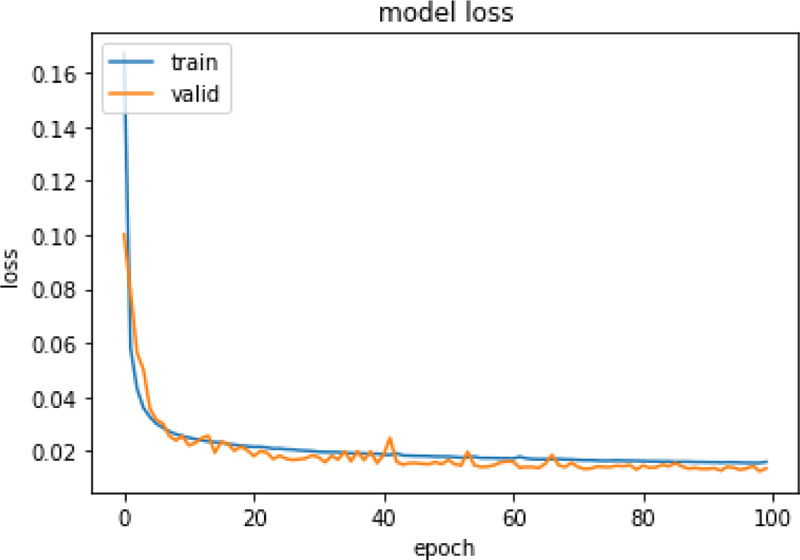
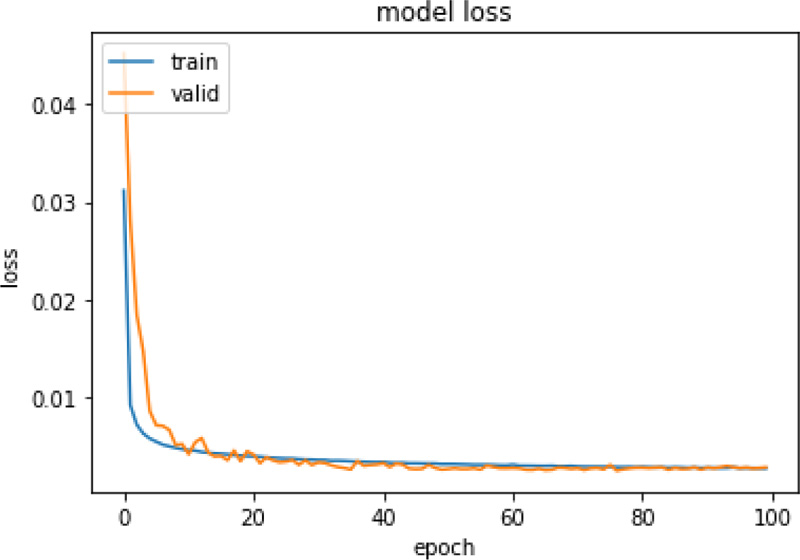
Same experiments were also run on X-ray images and Table 5 presents the performance of those models. For the CXR scan dataset, the autoencoder1 model with Kmeans performed better. Figs. (4 and 5) represent the training and validation loss graph of the best performing autoencoder models for both CT and CXR scan datasets. From the graphs, it can be observed that the training of both the models is stopped at 100 epochs as no significant loss is observed.
| Method | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|
| Autoencoder1 with Kmeans | 0.633 | 0.451 | 0.597 | 0.514 |
| Autoencoder2 with Kmeans | 0.664 | 0.484 | 0.535 | 0.508 |
| Autoencoder3 with Kmeans | 0.672 | 0.496 | 0.508 | 0.502 |
| Autoencoder4 with Kmeans | 0.668 | 0.494 | 0.997 | 0.661 |
| PCA with Kmeans | 0.351 | 0.225 | 0.409 | 0.290 |
| Method | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|
| Autoencoder1 with Kmeans | 0.664 | 0.709 | 0.709 | 0.709 |
| Autoencoder2 with Kmeans | 0.580 | 0.695 | 0.485 | 0.572 |
| Autoencoder3 with Kmeans | 0.571 | 0.580 | 0.932 | 0.715 |
| Autoencoder4 with Kmeans | 0.603 | 0.623 | 0.789 | 0.696 |
| PCA with Kmeans | 0.654 | 0.668 | 0.797 | 0.726 |
In the second set of experiments, PCA is used for feature extraction and dimensionality reduction, and those features are then fed into Kmeans for clustering. In Kmeans, the variance is set to 0.98 for both the CT and CXR scan datasets. The number of components is reduced from 4096 to 855 for the CT dataset and the number of components is reduced from 4096 to 788 for the CXR dataset. For both the CT and CXR scan datasets, the performance of PCA is poor when compared to the performance of autoencoders. The PCA based model achieved an accuracy of 35.1% and 65.4% for the CT and CXR dataset, respectively. Fig. (1) represents original and PCA retained images.
Pretrained CNN-based ImageNet models are widely used for many computer vision problems, including most medical imaging tasks. In this study, we have used VGG-16, VGG-19, and ResNet50 models by freezing all the pretrained layers for feature extraction, and clustering is performed by methods such as Kmeans and GMM. We have also incorporated PCA for dimensionality reduction and have reported the performance of the pretrained models with and without PCA in (Tables 6 and 7) for both CT and CXR scan datasets. The size of VGG-16, VGG-19, and ResNet50 features is 2048, 2048, and 8192, respectively for both CT and CXR datasets. We have not reduced the dimensionality of the VGG-16, VGG-19, and ResNet50 features in both CT and CXR datasets. Among these models, for the CT scan dataset, ResNet50-GMM without PCA performed better with an accuracy of 73.3%. The second-best performing model is VGG-19-GMM with PCA which achieved an accuracy of 71.8%. For the CXR scan dataset, the performance of models is poor and the maximum accuracy of 56.5% is achieved by both VGG16-Kmeans with and without PCA. For all GM experiments, we set n_components=2, covariance_type=’full’, tol=0.001, reg_covar=1e-06, max_iter=100, n_init=1, init_params=’kmeans’, weights_init=None, means_ init=None, precisions_init=None, random_state=None, and warm_start=False.
Table 6.
| Method | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|
| VGG16-KMeans (Without PCA) | 0.669 | 0.737 | 0.761 | 0.749 |
| VGG16-KMeans (With PCA) | 0.669 | 0.737 | 0.761 | 0.749 |
| VGG16-GMM (With PCA) | 0.669 | 0.748 | 0.755 | 0.751 |
| VGG19-KMeans (Without PCA) | 0.566 | 0.465 | 0.667 | 0.548 |
| VGG19-KMeans (With PCA) | 0.486 | 0.318 | 0.458 | 0.375 |
| VGG19-GMM (With PCA) | 0.718 | 0.667 | 0.918 | 0.772 |
| Resnet50-KMeans (Without PCA) | 0.479 | 0.335 | 0.443 | 0.381 |
| Resnet50-KMeans (With PCA) | 0.479 | 0.335 | 0.443 | 0.381 |
| Resnet50-GMM (With PCA) | 0.733 | 0.727 | 0.888 | 0.800 |
| Method | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|
| VGG16-KMeans (Without PCA) | 0.565 | 0.544 | 0.634 | 0.586 |
| VGG16-KMeans (With PCA) | 0.565 | 0.544 | 0.634 | 0.586 |
| VGG16-GMM (With PCA) | 0.561 | 0.535 | 0.626 | 0.577 |
| VGG19-KMeans (Without PCA) | 0.453 | 0.576 | 0.424 | 0.488 |
| VGG19-KMeans (With PCA) | 0.505 | 0.502 | 0.510 | 0.506 |
| VGG19-GMM (With PCA) | 0.517 | 0.487 | 0.536 | 0.510 |
| ResNet50-KMeans (Without PCA) | 0.403 | 0.425 | 0.319 | 0.364 |
| ResNet50-KMeans (With PCA) | 0.403 | 0.425 | 0.319 | 0.364 |
| ResNet50-GMM (With PCA) | 0.484 | 0.151 | 0.410 | 0.221 |
| Technique | Year | Method | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|---|---|
| Deep learning | Proposed | Autoencoder3-Resnet50 (Kmeans) | 0.679 | 0.503 | 0.977 | 0.664 |
| Deep learning | Proposed | Autoencoder3-Resnet50 (GMM) | 0.836 | 0.708 | 0.842 | 0.769 |
| Deep learning | 2017 | Autoencoder∗ (Kmeans)-Guo et al. [33] | 0.752 | 0.679 | 0.354 | 0.465 |
| Deep learning | 2017 | Autoencoder∗ (Kmeans)-Guo et al. [34] | 0.771 | 0.731 | 0.381 | 0.501 |
| Deep learning | 2018 | Autoencoder∗ (Kmeans∗) Aljalbout et al. [35] | 0.769 | 0.731 | 0.381 | 0.501 |
| Neural network | 2020 | Self-organizing map-King et al. [36] | 0.734 | 0.617 | 0.313 | 0.415 |
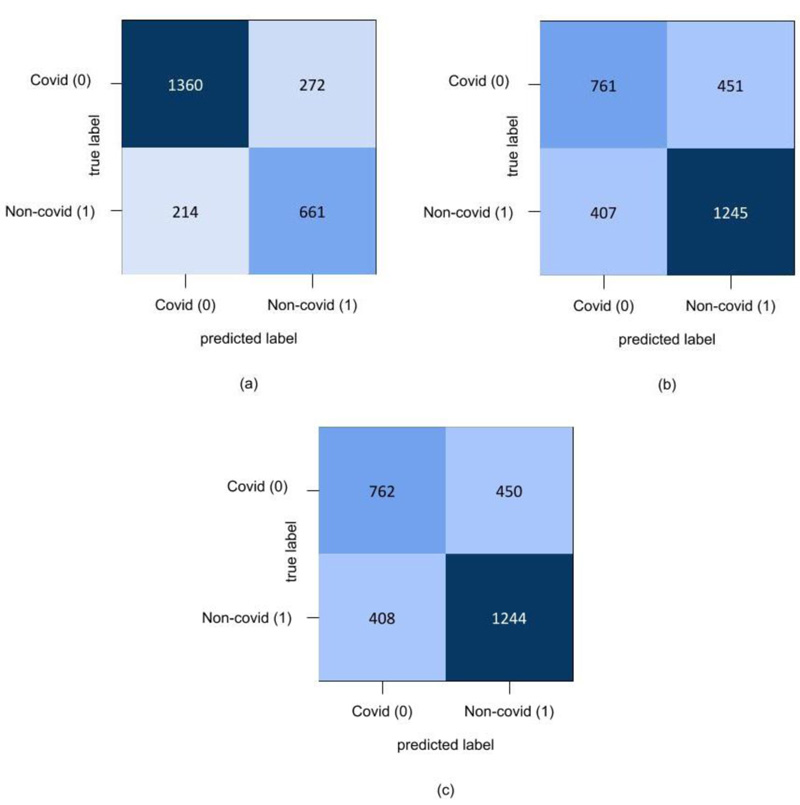
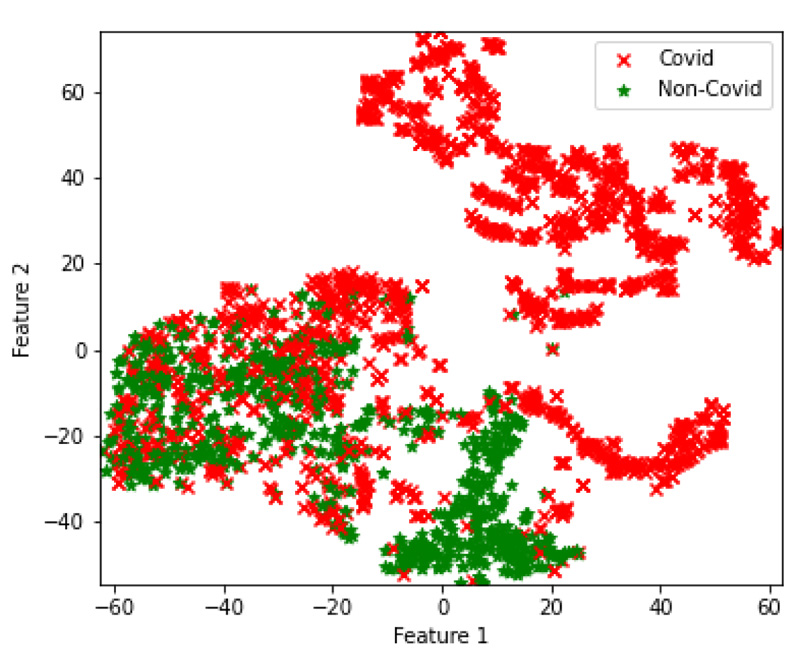
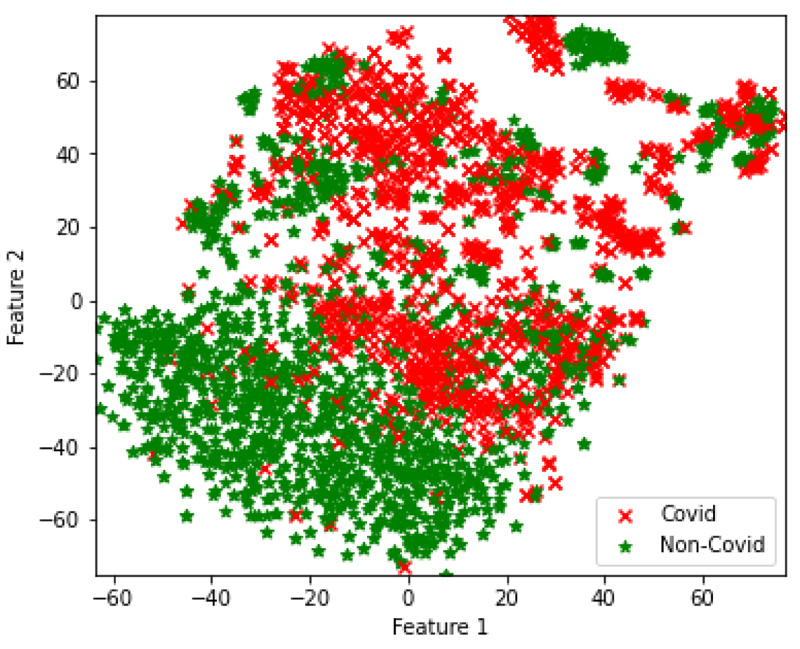
In the final set of experiments, four hybrid feature fusion-based models are trained and their performance is reported in Tables 8 and 9 for CT and CXR scan datasets, respectively. All four models have utilized PCA for feature extraction and data dimensionality reduction. For the CT scan dataset, two Autoencoder3-ResNet50 are trained; one with Kmeans and another one with GMM for final clustering. The number of features of Autoencoder3 and ResNet50 is 512 and 5638 respectively. These two feature sets are merged and passed into Kmeans and GMM. The Autocoder3-ResNet50 model with GMM performed better with an accuracy of 83.6% and Fig. (6A) presents its confusion matrix. Similarly, for the CXR scan dataset, Autoencoder1-VGG-16 models with GMM and Kmeans are trained, and both the models achieved an accuracy of 70%. Fig. (6b and 6c) present their confusion matrix. The number of features of Autoencoder1 and VGG-16 is 8192 and 2048, respectively. After we applied PCA on VGG-16 features, we retained all the features i.e. 2048 and finally, the features of Autoencoder1, VGG-16 with PCA and without PCA are merged and passed into Kmeans and GMM. The higher dimensional features from the best performing models for both datasets are transformed into lower dimensions and visualized using the t-SNE technique. Figs. (7 and 8) represent the t-SNE visualization of Autoencoder3-ResNet50 (CT scan) and Autoencoder1-VGG16 (CXR scan) models, respectively. The overlapping of the clusters can be seen in both the figures which shows the highly non-linear nature of the data. This is one of the reasons why GMM is preferred as it may produce better results.
Tables 8 and 9 show the performance of the proposed method with the other four well-known unsupervised methods. Out of four methods, a method based on self-organizing maps is proposed for COVID-19 detection. Other methods are evaluated on MNIST and other standard datasets in the original works. In our work, we have implemented those methods and evaluated the performance of COVID-19 detection using CT and X-ray datasets. In both the tables, the asterisk (*) with Autoencoder and Kmeans denotes that the methods are improved versions of Autoencoder and Kmeans, respectively. For both CT and X-ray datasets, the proposed method performed better than the existing four unsupervised methods.
| Technique | Year | Method | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|---|---|
| Deep learning | Proposed | Autoencoder1-VGG16 (KMeans) | 0.700 | 0.734 | 0.754 | 0.744 |
| Deep learning | Proposed | Autoencoder1-VGG16 (GMM) | 0.700 | 0.734 | 0.753 | 0.744 |
| Deep learning | 2017 | Autoencoder∗ (Kmeans)-Guo et al. [33] | 0.555 | 0.597 | 0.727 | 0.656 |
| Deep learning | 2017 | Autoencoder∗ (Kmeans)-Guo et al. [34] | 0.62 | 0.689 | 0.754 | 0.720 |
| Deep learning | 2018 | Autoencoder∗ (Kmeans∗) Aljalbout et al. [35] | 0.587 | 0.643 | 0.742 | 0.69 |
| Neural network | 2020 | Self-organizing map-King et al. [36] | 0.571 | 0.620 | 0.734 | 0.673 |
6. DISCUSSION AND FUTURE WORKS
Literature survey shows that there are many published papers on COVID-19 detection using CT and CXR dataset analysis using machine learning and deep learning. However, most of the published articles are based on supervised learning. These methods require labelled datasets. The availability of labelled datasets is less and often the labels are incomplete and inaccurate. The new CT and X-ray datasets have been added progressively to the existing datasets. Unsupervised learning is a type of machine learning which has been largely used in medical image analysis [38]. Unsupervised learning has the capability to learn the optimal features and cluster them into separate groups based on similar features without the labelled datasets. Following this work, various unsupervised learning approaches are applied and the detailed investigation and analysis of experiments for COVID-19 detection are shown. The proposed method performed better than the existing four unsupervised methods and showed an accuracy of 84% on the CT dataset and an accuracy of 70% CXR dataset. The major strength of the proposed work is unsupervised learning and the proposed method performance is shown on big datasets. Most of the published articles have used small datasets. Though the proposed method's performance is lesser compared to the existing supervised deep learning approaches, the proposed approach performed better than existing unsupervised learning methods. The proposed method may not be used as a tool for COVID-19 detection, however, the performance of the proposed method can be enhanced by enhancing the proposed approach to correct misclassifications.
In the current work, the CT scan and CXR datasets are used for COVID-19 detection. In the future, we are planning to add additional features for each patient such as symptoms, age, gender, etc. Finding the correlation between these features, various conditions and infection will be an important direction toward COVID-19 detection in the current pandemic disease. Besides, to cluster the datasets as just COVID-19 and Non-COVID-19, we are planning to group data samples into more clusters such as early-stage, final-stage, other diseases, etc. This type of feature helps doctors to track various diseases.
In the current work, we have used many unsupervised methods such as convolutional autoencoders, PCA-based models, pretrained CNN ImageNet architectures for feature extraction. Besides, we have used a simple feature fusion approach which showed better performances compared to the non-feature fusion methods. However, instead of simply merging the features from different unsupervised models, information fusion methodologies can be employed [39]. This type of method can improve existing performances. For clustering purposes, Kmeans and GMM are used. There are many advanced clustering methods proposed by others recently which can enhance the performance of Kmeans and GMM [38]. This type of work can be considered another significant direction toward future work [40, 41].
CONCLUSION
In this work, we have used convolutional autoencoder networks, PCA-based models, and pretrained CNN ImageNet architectures for feature extraction and we have applied methods such as Kmeans and GMM for clustering the CT and CXR scan COVID-19 images. We have also proposed a hybrid deep learning model based on feature fusion that combines the features learned by the best performing convolutional autoencoder and pretrained CNN models. The feature fusion-based approach has significantly improved the performance of other models with an accuracy of 83%. We have visualized the features using the t-SNE technique. Even though there is a significant enhancement in the performance of the proposed model, its accuracy seems lower when compared to the supervised learning-based models. Therefore, in the future, more experiments can be conducted incorporating advanced feature fusion methods for improving the performance further.
LIST OF ABBREVIATIONS
| CXR | = Chest X-ray |
| CT | = Computed Tomography |
| PCA | = Principal Component Analysis |
| TP | = True Positive |
| TN | = True Negative |
| FN | = False Negative |
| FP | = False Positive |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author, Vinayakumar Ravi, on special request.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


